Model info
| Transcription factor | Bhlhe40 | ||||||||
| Model | BHE40_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 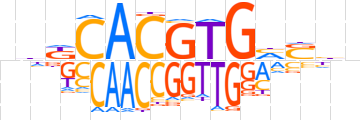 | ||||||||
| LOGO (reverse complement) | 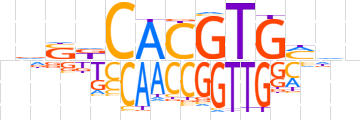 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vhKCACGTGhShn | ||||||||
| Best auROC (human) | 0.84 | ||||||||
| Best auROC (mouse) | 0.971 | ||||||||
| Peak sets in benchmark (human) | 10 | ||||||||
| Peak sets in benchmark (mouse) | 5 | ||||||||
| Aligned words | 339 | ||||||||
| TF family | Hairy-related factors {1.2.4} | ||||||||
| TF subfamily | Hairy-like factors {1.2.4.1} | ||||||||
| MGI | MGI:1097714 | ||||||||
| EntrezGene | GeneID:20893 (SSTAR profile) | ||||||||
| UniProt ID | BHE40_MOUSE | ||||||||
| UniProt AC | O35185 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Bhlhe40 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 18.0 | 9.0 | 7.0 | 26.0 | 50.0 | 23.0 | 11.0 | 41.0 | 29.0 | 25.0 | 11.0 | 32.0 | 4.0 | 6.0 | 19.0 | 23.0 |
| 02 | 13.0 | 5.0 | 61.0 | 22.0 | 10.0 | 1.0 | 16.0 | 36.0 | 3.0 | 3.0 | 30.0 | 12.0 | 12.0 | 6.0 | 80.0 | 24.0 |
| 03 | 19.0 | 18.0 | 1.0 | 0.0 | 0.0 | 15.0 | 0.0 | 0.0 | 11.0 | 174.0 | 1.0 | 1.0 | 0.0 | 93.0 | 1.0 | 0.0 |
| 04 | 30.0 | 0.0 | 0.0 | 0.0 | 296.0 | 1.0 | 1.0 | 2.0 | 2.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 05 | 0.0 | 303.0 | 14.0 | 12.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 20.0 | 18.0 | 270.0 | 0.0 | 0.0 | 2.0 | 12.0 | 0.0 | 0.0 | 0.0 | 12.0 | 0.0 |
| 07 | 1.0 | 0.0 | 0.0 | 19.0 | 1.0 | 0.0 | 1.0 | 18.0 | 22.0 | 7.0 | 5.0 | 260.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 24.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 5.0 | 1.0 | 0.0 | 0.0 | 295.0 | 2.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 175.0 | 99.0 | 18.0 | 39.0 | 1.0 | 0.0 | 0.0 | 2.0 |
| 10 | 4.0 | 119.0 | 40.0 | 13.0 | 19.0 | 62.0 | 12.0 | 6.0 | 1.0 | 10.0 | 7.0 | 0.0 | 2.0 | 20.0 | 16.0 | 3.0 |
| 11 | 0.0 | 17.0 | 4.0 | 5.0 | 43.0 | 69.0 | 16.0 | 83.0 | 18.0 | 22.0 | 9.0 | 26.0 | 5.0 | 6.0 | 7.0 | 4.0 |
| 12 | 7.0 | 14.0 | 30.0 | 15.0 | 35.0 | 39.0 | 13.0 | 27.0 | 6.0 | 9.0 | 10.0 | 11.0 | 9.0 | 30.0 | 55.0 | 24.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.145 | -0.819 | -1.059 | 0.216 | 0.863 | 0.095 | -0.625 | 0.667 | 0.324 | 0.177 | -0.625 | 0.421 | -1.583 | -1.205 | -0.092 | 0.095 |
| 02 | -0.463 | -1.376 | 1.061 | 0.052 | -0.718 | -2.746 | -0.261 | 0.538 | -1.843 | -1.843 | 0.357 | -0.541 | -0.541 | -1.205 | 1.331 | 0.137 |
| 03 | -0.092 | -0.145 | -2.746 | -4.069 | -4.069 | -0.324 | -4.069 | -4.069 | -0.625 | 2.105 | -2.746 | -2.746 | -4.069 | 1.481 | -2.746 | -4.069 |
| 04 | 0.357 | -4.069 | -4.069 | -4.069 | 2.636 | -2.746 | -2.746 | -2.196 | -2.196 | -4.069 | -4.069 | -2.746 | -2.746 | -4.069 | -4.069 | -4.069 |
| 05 | -4.069 | 2.659 | -0.391 | -0.541 | -4.069 | -2.746 | -4.069 | -4.069 | -4.069 | -2.746 | -4.069 | -4.069 | -4.069 | -1.843 | -4.069 | -4.069 |
| 06 | -4.069 | -4.069 | -4.069 | -4.069 | -0.042 | -0.145 | 2.544 | -4.069 | -4.069 | -2.196 | -0.541 | -4.069 | -4.069 | -4.069 | -0.541 | -4.069 |
| 07 | -2.746 | -4.069 | -4.069 | -0.092 | -2.746 | -4.069 | -2.746 | -0.145 | 0.052 | -1.059 | -1.376 | 2.506 | -4.069 | -4.069 | -4.069 | -4.069 |
| 08 | -4.069 | -4.069 | 0.137 | -4.069 | -4.069 | -4.069 | -1.059 | -4.069 | -4.069 | -4.069 | -1.376 | -2.746 | -4.069 | -4.069 | 2.632 | -2.196 |
| 09 | -4.069 | -4.069 | -4.069 | -4.069 | -4.069 | -4.069 | -4.069 | -4.069 | 2.111 | 1.543 | -0.145 | 0.617 | -2.746 | -4.069 | -4.069 | -2.196 |
| 10 | -1.583 | 1.726 | 0.642 | -0.463 | -0.092 | 1.077 | -0.541 | -1.205 | -2.746 | -0.718 | -1.059 | -4.069 | -2.196 | -0.042 | -0.261 | -1.843 |
| 11 | -4.069 | -0.201 | -1.583 | -1.376 | 0.714 | 1.184 | -0.261 | 1.367 | -0.145 | 0.052 | -0.819 | 0.216 | -1.376 | -1.205 | -1.059 | -1.583 |
| 12 | -1.059 | -0.391 | 0.357 | -0.324 | 0.51 | 0.617 | -0.463 | 0.253 | -1.205 | -0.819 | -0.718 | -0.625 | -0.819 | 0.357 | 0.958 | 0.137 |