Model info
| Transcription factor | Nr2f2 | ||||||||
| Model | COT2_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 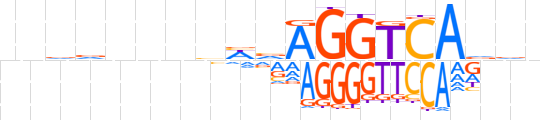 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnddnnnhhvAGGTCAdvn | ||||||||
| Best auROC (human) | 0.95 | ||||||||
| Best auROC (mouse) | 0.874 | ||||||||
| Peak sets in benchmark (human) | 28 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | RXR-related receptors (NR2) {2.1.3} | ||||||||
| TF subfamily | COUP-like receptors (NR2F) {2.1.3.5} | ||||||||
| MGI | MGI:1352452 | ||||||||
| EntrezGene | GeneID:11819 (SSTAR profile) | ||||||||
| UniProt ID | COT2_MOUSE | ||||||||
| UniProt AC | P43135 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Nr2f2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 45.222 | 19.877 | 21.67 | 20.122 | 64.444 | 11.544 | 1.027 | 43.412 | 31.767 | 25.349 | 39.2 | 32.982 | 34.372 | 26.162 | 43.843 | 39.008 |
| 02 | 63.22 | 21.379 | 68.076 | 23.13 | 51.489 | 8.931 | 7.731 | 14.781 | 32.12 | 12.296 | 42.914 | 18.41 | 32.775 | 16.804 | 54.529 | 31.416 |
| 03 | 80.841 | 13.722 | 62.58 | 22.461 | 40.487 | 6.188 | 2.868 | 9.867 | 50.039 | 30.0 | 72.617 | 20.594 | 29.367 | 9.8 | 24.986 | 23.585 |
| 04 | 62.789 | 13.813 | 79.702 | 44.43 | 21.399 | 14.147 | 2.124 | 22.038 | 42.69 | 30.799 | 74.225 | 15.335 | 8.625 | 26.837 | 16.728 | 24.317 |
| 05 | 24.992 | 32.756 | 50.778 | 26.978 | 37.106 | 25.639 | 3.331 | 19.521 | 36.319 | 10.504 | 91.485 | 34.471 | 24.004 | 16.123 | 29.43 | 36.565 |
| 06 | 21.669 | 34.908 | 33.33 | 32.513 | 17.001 | 26.355 | 3.105 | 38.56 | 37.319 | 40.846 | 27.496 | 69.363 | 8.226 | 36.16 | 29.006 | 44.143 |
| 07 | 23.745 | 24.995 | 22.569 | 12.905 | 37.133 | 58.419 | 4.904 | 37.813 | 21.454 | 33.333 | 16.636 | 21.514 | 24.01 | 91.949 | 35.534 | 33.086 |
| 08 | 55.037 | 17.779 | 18.664 | 14.861 | 145.166 | 23.956 | 7.99 | 31.585 | 48.006 | 19.768 | 5.324 | 6.546 | 42.995 | 9.601 | 19.716 | 33.006 |
| 09 | 135.56 | 50.889 | 92.283 | 12.472 | 44.499 | 13.488 | 2.52 | 10.597 | 19.55 | 10.084 | 21.149 | 0.911 | 28.437 | 10.015 | 35.021 | 12.526 |
| 10 | 192.931 | 0.0 | 35.115 | 0.0 | 78.616 | 0.0 | 3.055 | 2.804 | 129.945 | 0.0 | 21.027 | 0.0 | 17.121 | 0.0 | 19.384 | 0.0 |
| 11 | 1.813 | 0.0 | 398.936 | 17.864 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 78.582 | 0.0 | 0.0 | 0.0 | 2.804 | 0.0 |
| 12 | 0.0 | 0.0 | 1.813 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.019 | 0.0 | 434.938 | 40.366 | 0.0 | 0.0 | 15.275 | 2.589 |
| 13 | 0.0 | 0.0 | 0.0 | 5.019 | 0.0 | 0.0 | 0.0 | 0.0 | 1.072 | 5.892 | 68.504 | 376.558 | 0.0 | 0.783 | 1.002 | 41.17 |
| 14 | 0.0 | 1.072 | 0.0 | 0.0 | 0.0 | 6.675 | 0.0 | 0.0 | 0.0 | 65.631 | 0.808 | 3.067 | 0.0 | 380.088 | 23.762 | 18.897 |
| 15 | 0.0 | 0.0 | 0.0 | 0.0 | 449.095 | 0.0 | 3.539 | 0.832 | 22.448 | 0.0 | 0.0 | 2.122 | 19.392 | 0.0 | 2.572 | 0.0 |
| 16 | 138.134 | 71.062 | 204.395 | 77.343 | 0.0 | 0.0 | 0.0 | 0.0 | 3.786 | 0.832 | 0.0 | 1.493 | 0.832 | 0.0 | 0.944 | 1.178 |
| 17 | 57.492 | 23.731 | 51.469 | 10.06 | 40.617 | 18.28 | 3.107 | 9.89 | 74.97 | 33.452 | 75.613 | 21.304 | 4.931 | 27.674 | 28.389 | 19.02 |
| 18 | 35.987 | 12.662 | 78.881 | 50.479 | 32.299 | 32.46 | 3.706 | 34.673 | 55.09 | 21.185 | 48.127 | 34.177 | 6.838 | 6.467 | 24.433 | 22.536 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.366 | -0.445 | -0.361 | -0.433 | 0.717 | -0.975 | -3.107 | 0.325 | 0.016 | -0.206 | 0.224 | 0.053 | 0.094 | -0.175 | 0.335 | 0.219 |
| 02 | 0.698 | -0.374 | 0.772 | -0.297 | 0.495 | -1.222 | -1.36 | -0.735 | 0.027 | -0.914 | 0.314 | -0.521 | 0.047 | -0.61 | 0.551 | 0.005 |
| 03 | 0.943 | -0.807 | 0.688 | -0.325 | 0.256 | -1.571 | -2.274 | -1.127 | 0.466 | -0.04 | 0.836 | -0.411 | -0.061 | -1.133 | -0.221 | -0.277 |
| 04 | 0.692 | -0.801 | 0.929 | 0.348 | -0.373 | -0.778 | -2.533 | -0.344 | 0.309 | -0.014 | 0.858 | -0.699 | -1.256 | -0.15 | -0.614 | -0.247 |
| 05 | -0.22 | 0.046 | 0.481 | -0.145 | 0.17 | -0.195 | -2.141 | -0.463 | 0.149 | -1.066 | 1.066 | 0.097 | -0.26 | -0.65 | -0.059 | 0.155 |
| 06 | -0.361 | 0.109 | 0.064 | 0.039 | -0.598 | -0.168 | -2.203 | 0.208 | 0.175 | 0.265 | -0.126 | 0.791 | -1.301 | 0.144 | -0.074 | 0.342 |
| 07 | -0.271 | -0.22 | -0.321 | -0.867 | 0.171 | 0.62 | -1.788 | 0.189 | -0.371 | 0.064 | -0.62 | -0.368 | -0.26 | 1.071 | 0.127 | 0.056 |
| 08 | 0.561 | -0.555 | -0.507 | -0.73 | 1.526 | -0.262 | -1.329 | 0.011 | 0.425 | -0.451 | -1.712 | -1.518 | 0.316 | -1.153 | -0.453 | 0.054 |
| 09 | 1.458 | 0.483 | 1.075 | -0.9 | 0.35 | -0.824 | -2.387 | -1.058 | -0.462 | -1.106 | -0.385 | -3.192 | -0.093 | -1.112 | 0.113 | -0.896 |
| 10 | 1.81 | -4.4 | 0.115 | -4.4 | 0.915 | -4.4 | -2.218 | -2.293 | 1.416 | -4.4 | -0.39 | -4.4 | -0.592 | -4.4 | -0.47 | -4.4 |
| 11 | -2.665 | -4.4 | 2.535 | -0.55 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 0.915 | -4.4 | -4.4 | -4.4 | -2.293 | -4.4 |
| 12 | -4.4 | -4.4 | -2.665 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -1.767 | -4.4 | 2.622 | 0.253 | -4.4 | -4.4 | -0.703 | -2.363 |
| 13 | -4.4 | -4.4 | -4.4 | -1.767 | -4.4 | -4.4 | -4.4 | -4.4 | -3.076 | -1.617 | 0.778 | 2.478 | -4.4 | -3.296 | -3.125 | 0.273 |
| 14 | -4.4 | -3.076 | -4.4 | -4.4 | -4.4 | -1.499 | -4.4 | -4.4 | -4.4 | 0.736 | -3.275 | -2.214 | -4.4 | 2.487 | -0.27 | -0.495 |
| 15 | -4.4 | -4.4 | -4.4 | -4.4 | 2.654 | -4.4 | -2.086 | -3.255 | -0.326 | -4.4 | -4.4 | -2.534 | -0.47 | -4.4 | -2.369 | -4.4 |
| 16 | 1.477 | 0.815 | 1.868 | 0.899 | -4.4 | -4.4 | -4.4 | -4.4 | -2.025 | -3.255 | -4.4 | -2.822 | -3.255 | -4.4 | -3.167 | -3.006 |
| 17 | 0.604 | -0.271 | 0.494 | -1.108 | 0.259 | -0.528 | -2.203 | -1.124 | 0.868 | 0.067 | 0.876 | -0.377 | -1.783 | -0.12 | -0.095 | -0.489 |
| 18 | 0.14 | -0.886 | 0.918 | 0.475 | 0.033 | 0.038 | -2.045 | 0.103 | 0.562 | -0.383 | 0.428 | 0.088 | -1.477 | -1.529 | -0.243 | -0.322 |