Model info
| Transcription factor | Dmrtb1 | ||||||||
| Model | DMRTB_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 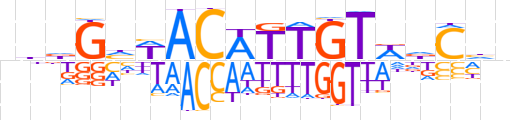 | ||||||||
| LOGO (reverse complement) | 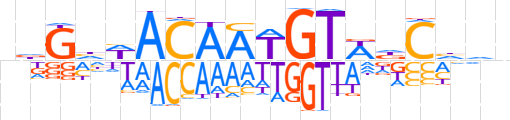 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbdGhWACATTGTWdChn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.9 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 460 | ||||||||
| TF family | DMRT {2.5.1} | ||||||||
| TF subfamily | DMRTB1 {2.5.1.0.6} | ||||||||
| MGI | MGI:1927125 | ||||||||
| EntrezGene | GeneID:56296 (SSTAR profile) | ||||||||
| UniProt ID | DMRTB_MOUSE | ||||||||
| UniProt AC | A2A9I7 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Dmrtb1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 17.0 | 28.0 | 41.0 | 37.0 | 12.0 | 25.0 | 6.0 | 65.0 | 11.0 | 18.0 | 15.0 | 30.0 | 8.0 | 25.0 | 36.0 | 42.0 |
| 02 | 11.0 | 2.0 | 19.0 | 16.0 | 28.0 | 11.0 | 4.0 | 53.0 | 22.0 | 7.0 | 30.0 | 39.0 | 19.0 | 6.0 | 75.0 | 74.0 |
| 03 | 2.0 | 1.0 | 75.0 | 2.0 | 2.0 | 3.0 | 19.0 | 2.0 | 9.0 | 0.0 | 113.0 | 6.0 | 2.0 | 7.0 | 167.0 | 6.0 |
| 04 | 11.0 | 2.0 | 0.0 | 2.0 | 1.0 | 8.0 | 0.0 | 2.0 | 124.0 | 153.0 | 8.0 | 89.0 | 3.0 | 4.0 | 2.0 | 7.0 |
| 05 | 25.0 | 18.0 | 2.0 | 94.0 | 53.0 | 14.0 | 1.0 | 99.0 | 6.0 | 2.0 | 2.0 | 0.0 | 19.0 | 7.0 | 0.0 | 74.0 |
| 06 | 100.0 | 1.0 | 0.0 | 2.0 | 37.0 | 1.0 | 3.0 | 0.0 | 4.0 | 0.0 | 0.0 | 1.0 | 256.0 | 1.0 | 0.0 | 10.0 |
| 07 | 1.0 | 386.0 | 4.0 | 6.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 1.0 | 9.0 | 0.0 | 3.0 |
| 08 | 1.0 | 0.0 | 0.0 | 1.0 | 296.0 | 5.0 | 3.0 | 97.0 | 0.0 | 0.0 | 0.0 | 4.0 | 2.0 | 1.0 | 0.0 | 6.0 |
| 09 | 0.0 | 0.0 | 73.0 | 226.0 | 0.0 | 0.0 | 1.0 | 5.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 21.0 | 87.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.0 | 2.0 | 1.0 | 80.0 | 43.0 | 3.0 | 4.0 | 271.0 |
| 11 | 7.0 | 3.0 | 44.0 | 1.0 | 0.0 | 2.0 | 2.0 | 1.0 | 0.0 | 1.0 | 4.0 | 0.0 | 3.0 | 7.0 | 326.0 | 15.0 |
| 12 | 2.0 | 0.0 | 0.0 | 8.0 | 3.0 | 0.0 | 1.0 | 9.0 | 2.0 | 3.0 | 3.0 | 368.0 | 2.0 | 2.0 | 2.0 | 11.0 |
| 13 | 6.0 | 1.0 | 0.0 | 2.0 | 4.0 | 0.0 | 0.0 | 1.0 | 1.0 | 2.0 | 0.0 | 3.0 | 226.0 | 18.0 | 30.0 | 122.0 |
| 14 | 64.0 | 8.0 | 67.0 | 98.0 | 3.0 | 3.0 | 3.0 | 12.0 | 3.0 | 7.0 | 6.0 | 14.0 | 32.0 | 3.0 | 53.0 | 40.0 |
| 15 | 7.0 | 83.0 | 5.0 | 7.0 | 9.0 | 10.0 | 0.0 | 2.0 | 8.0 | 117.0 | 2.0 | 2.0 | 11.0 | 145.0 | 1.0 | 7.0 |
| 16 | 10.0 | 14.0 | 4.0 | 7.0 | 127.0 | 120.0 | 21.0 | 87.0 | 4.0 | 2.0 | 1.0 | 1.0 | 4.0 | 7.0 | 1.0 | 6.0 |
| 17 | 50.0 | 31.0 | 46.0 | 18.0 | 74.0 | 26.0 | 5.0 | 38.0 | 9.0 | 8.0 | 6.0 | 4.0 | 17.0 | 24.0 | 34.0 | 26.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.417 | 0.073 | 0.45 | 0.349 | -0.757 | -0.039 | -1.42 | 0.908 | -0.841 | -0.361 | -0.54 | 0.141 | -1.147 | -0.039 | 0.321 | 0.474 |
| 02 | -0.841 | -2.407 | -0.308 | -0.477 | 0.073 | -0.841 | -1.796 | 0.705 | -0.164 | -1.274 | 0.141 | 0.401 | -0.308 | -1.42 | 1.05 | 1.037 |
| 03 | -2.407 | -2.953 | 1.05 | -2.407 | -2.407 | -2.056 | -0.308 | -2.407 | -1.034 | -4.248 | 1.458 | -1.42 | -2.407 | -1.274 | 1.848 | -1.42 |
| 04 | -0.841 | -2.407 | -4.248 | -2.407 | -2.953 | -1.147 | -4.248 | -2.407 | 1.551 | 1.76 | -1.147 | 1.22 | -2.056 | -1.796 | -2.407 | -1.274 |
| 05 | -0.039 | -0.361 | -2.407 | 1.275 | 0.705 | -0.607 | -2.953 | 1.326 | -1.42 | -2.407 | -2.407 | -4.248 | -0.308 | -1.274 | -4.248 | 1.037 |
| 06 | 1.336 | -2.953 | -4.248 | -2.407 | 0.349 | -2.953 | -2.056 | -4.248 | -1.796 | -4.248 | -4.248 | -2.953 | 2.274 | -2.953 | -4.248 | -0.933 |
| 07 | -2.953 | 2.684 | -1.796 | -1.42 | -4.248 | -2.056 | -4.248 | -4.248 | -4.248 | -2.056 | -4.248 | -4.248 | -2.953 | -1.034 | -4.248 | -2.056 |
| 08 | -2.953 | -4.248 | -4.248 | -2.953 | 2.419 | -1.59 | -2.056 | 1.306 | -4.248 | -4.248 | -4.248 | -1.796 | -2.407 | -2.953 | -4.248 | -1.42 |
| 09 | -4.248 | -4.248 | 1.023 | 2.15 | -4.248 | -4.248 | -2.953 | -1.59 | -4.248 | -4.248 | -4.248 | -2.056 | -4.248 | -4.248 | -0.21 | 1.198 |
| 10 | -4.248 | -4.248 | -4.248 | -4.248 | -4.248 | -4.248 | -4.248 | -4.248 | -0.757 | -2.407 | -2.953 | 1.114 | 0.497 | -2.056 | -1.796 | 2.331 |
| 11 | -1.274 | -2.056 | 0.52 | -2.953 | -4.248 | -2.407 | -2.407 | -2.953 | -4.248 | -2.953 | -1.796 | -4.248 | -2.056 | -1.274 | 2.516 | -0.54 |
| 12 | -2.407 | -4.248 | -4.248 | -1.147 | -2.056 | -4.248 | -2.953 | -1.034 | -2.407 | -2.056 | -2.056 | 2.637 | -2.407 | -2.407 | -2.407 | -0.841 |
| 13 | -1.42 | -2.953 | -4.248 | -2.407 | -1.796 | -4.248 | -4.248 | -2.953 | -2.953 | -2.407 | -4.248 | -2.056 | 2.15 | -0.361 | 0.141 | 1.535 |
| 14 | 0.892 | -1.147 | 0.938 | 1.316 | -2.056 | -2.056 | -2.056 | -0.757 | -2.056 | -1.274 | -1.42 | -0.607 | 0.205 | -2.056 | 0.705 | 0.426 |
| 15 | -1.274 | 1.151 | -1.59 | -1.274 | -1.034 | -0.933 | -4.248 | -2.407 | -1.147 | 1.493 | -2.407 | -2.407 | -0.841 | 1.707 | -2.953 | -1.274 |
| 16 | -0.933 | -0.607 | -1.796 | -1.274 | 1.575 | 1.518 | -0.21 | 1.198 | -1.796 | -2.407 | -2.953 | -2.953 | -1.796 | -1.274 | -2.953 | -1.42 |
| 17 | 0.647 | 0.174 | 0.564 | -0.361 | 1.037 | 0.0 | -1.59 | 0.375 | -1.034 | -1.147 | -1.42 | -1.796 | -0.417 | -0.079 | 0.265 | 0.0 |