Model info
| Transcription factor | Esrrb | ||||||||
| Model | ERR2_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 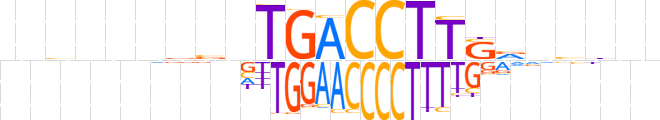 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nndbnbSAAGGTCAnvbnvnvnn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.994 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 14 | ||||||||
| Aligned words | 529 | ||||||||
| TF family | Steroid hormone receptors (NR3) {2.1.1} | ||||||||
| TF subfamily | ER-like receptors (NR3A&B) {2.1.1.2} | ||||||||
| MGI | MGI:1346832 | ||||||||
| EntrezGene | GeneID:26380 (SSTAR profile) | ||||||||
| UniProt ID | ERR2_MOUSE | ||||||||
| UniProt AC | Q61539 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Esrrb expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 30.217 | 29.133 | 43.738 | 13.331 | 38.776 | 32.933 | 3.872 | 44.62 | 63.312 | 19.787 | 49.554 | 19.721 | 9.939 | 31.624 | 43.154 | 26.29 |
| 02 | 82.016 | 17.025 | 29.849 | 13.354 | 38.335 | 32.095 | 11.854 | 31.194 | 72.756 | 19.012 | 32.849 | 15.7 | 11.172 | 26.589 | 24.637 | 41.563 |
| 03 | 26.038 | 26.715 | 134.793 | 16.733 | 33.227 | 23.895 | 13.669 | 23.93 | 13.985 | 32.161 | 37.329 | 15.714 | 6.21 | 24.003 | 39.799 | 31.798 |
| 04 | 10.133 | 21.102 | 37.337 | 10.889 | 23.985 | 35.093 | 7.839 | 39.856 | 26.053 | 36.164 | 59.319 | 104.054 | 1.941 | 28.987 | 46.044 | 11.202 |
| 05 | 8.8 | 14.128 | 20.97 | 18.214 | 7.905 | 48.694 | 3.082 | 61.667 | 25.538 | 33.437 | 28.22 | 63.345 | 1.91 | 28.809 | 12.839 | 122.443 |
| 06 | 10.472 | 21.89 | 10.816 | 0.974 | 25.134 | 91.119 | 1.921 | 6.894 | 3.932 | 39.166 | 20.006 | 2.006 | 13.237 | 210.16 | 38.32 | 3.952 |
| 07 | 21.367 | 0.0 | 31.408 | 0.0 | 355.407 | 0.955 | 3.041 | 2.933 | 61.898 | 0.996 | 7.179 | 0.99 | 7.906 | 0.0 | 5.921 | 0.0 |
| 08 | 444.709 | 0.0 | 1.869 | 0.0 | 1.951 | 0.0 | 0.0 | 0.0 | 45.558 | 0.0 | 1.99 | 0.0 | 3.923 | 0.0 | 0.0 | 0.0 |
| 09 | 2.19 | 0.0 | 493.95 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.86 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 2.19 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.029 | 0.0 | 495.749 | 1.032 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 0.0 | 0.0 | 0.0 | 1.029 | 0.0 | 0.0 | 0.0 | 0.0 | 7.946 | 0.983 | 34.853 | 454.157 | 0.0 | 0.0 | 0.0 | 1.032 |
| 12 | 0.0 | 7.946 | 0.0 | 0.0 | 0.0 | 0.983 | 0.0 | 0.0 | 0.0 | 34.853 | 0.0 | 0.0 | 0.959 | 434.718 | 12.577 | 7.964 |
| 13 | 0.959 | 0.0 | 0.0 | 0.0 | 467.585 | 0.0 | 10.916 | 0.0 | 11.606 | 0.0 | 0.972 | 0.0 | 7.964 | 0.0 | 0.0 | 0.0 |
| 14 | 86.902 | 174.774 | 114.49 | 111.947 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.888 | 0.989 | 5.01 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 14.598 | 14.609 | 54.811 | 2.884 | 41.079 | 91.076 | 27.034 | 21.473 | 11.105 | 37.927 | 54.561 | 11.886 | 8.923 | 43.329 | 40.97 | 23.735 |
| 16 | 6.778 | 19.167 | 44.711 | 5.049 | 11.891 | 98.98 | 10.834 | 65.236 | 37.313 | 58.052 | 58.728 | 23.283 | 4.193 | 21.004 | 22.991 | 11.791 |
| 17 | 18.575 | 19.782 | 18.781 | 3.037 | 58.714 | 32.46 | 5.149 | 100.879 | 31.842 | 30.337 | 40.041 | 35.043 | 9.235 | 27.533 | 27.985 | 40.606 |
| 18 | 43.699 | 12.9 | 52.646 | 9.12 | 36.394 | 40.152 | 12.952 | 20.614 | 28.358 | 25.46 | 28.127 | 10.011 | 13.731 | 65.295 | 80.52 | 20.02 |
| 19 | 37.742 | 18.766 | 52.712 | 12.963 | 57.02 | 32.834 | 7.938 | 46.016 | 41.508 | 42.251 | 71.478 | 19.008 | 7.683 | 15.394 | 30.797 | 5.891 |
| 20 | 25.828 | 30.21 | 80.01 | 7.904 | 39.538 | 36.766 | 14.804 | 18.136 | 27.782 | 30.712 | 70.789 | 33.641 | 13.93 | 16.942 | 41.148 | 11.859 |
| 21 | 26.212 | 28.436 | 43.221 | 9.209 | 42.006 | 44.758 | 4.12 | 23.746 | 40.55 | 59.643 | 67.359 | 39.2 | 9.892 | 22.687 | 23.171 | 15.79 |
| 22 | 23.763 | 38.243 | 43.616 | 13.038 | 43.584 | 27.715 | 4.996 | 79.229 | 34.368 | 23.929 | 46.08 | 33.495 | 10.062 | 32.377 | 18.67 | 26.835 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.033 | -0.069 | 0.333 | -0.836 | 0.213 | 0.052 | -2.005 | 0.352 | 0.7 | -0.45 | 0.457 | -0.453 | -1.12 | 0.012 | 0.319 | -0.171 |
| 02 | 0.957 | -0.597 | -0.045 | -0.834 | 0.202 | 0.026 | -0.949 | -0.002 | 0.838 | -0.489 | 0.049 | -0.676 | -1.007 | -0.159 | -0.234 | 0.282 |
| 03 | -0.18 | -0.155 | 1.452 | -0.614 | 0.061 | -0.265 | -0.811 | -0.263 | -0.789 | 0.028 | 0.176 | -0.675 | -1.567 | -0.26 | 0.239 | 0.017 |
| 04 | -1.101 | -0.387 | 0.176 | -1.032 | -0.261 | 0.115 | -1.347 | 0.241 | -0.179 | 0.144 | 0.635 | 1.194 | -2.609 | -0.074 | 0.384 | -1.004 |
| 05 | -1.236 | -0.779 | -0.393 | -0.531 | -1.339 | 0.439 | -2.21 | 0.674 | -0.199 | 0.067 | -0.101 | 0.7 | -2.622 | -0.08 | -0.872 | 1.356 |
| 06 | -1.069 | -0.351 | -1.038 | -3.145 | -0.215 | 1.062 | -2.617 | -1.469 | -1.991 | 0.223 | -0.439 | -2.581 | -0.842 | 1.895 | 0.202 | -1.986 |
| 07 | -0.374 | -4.4 | 0.005 | -4.4 | 2.42 | -3.159 | -2.222 | -2.254 | 0.677 | -3.129 | -1.431 | -3.133 | -1.339 | -4.4 | -1.612 | -4.4 |
| 08 | 2.644 | -4.4 | -2.64 | -4.4 | -2.605 | -4.4 | -4.4 | -4.4 | 0.373 | -4.4 | -2.588 | -4.4 | -1.993 | -4.4 | -4.4 | -4.4 |
| 09 | -2.507 | -4.4 | 2.749 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.008 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 10 | -4.4 | -4.4 | -2.507 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.106 | -4.4 | 2.752 | -3.103 | -4.4 | -4.4 | -4.4 | -4.4 |
| 11 | -4.4 | -4.4 | -4.4 | -3.106 | -4.4 | -4.4 | -4.4 | -4.4 | -1.334 | -3.138 | 0.108 | 2.665 | -4.4 | -4.4 | -4.4 | -3.103 |
| 12 | -4.4 | -1.334 | -4.4 | -4.4 | -4.4 | -3.138 | -4.4 | -4.4 | -4.4 | 0.108 | -4.4 | -4.4 | -3.156 | 2.621 | -0.892 | -1.332 |
| 13 | -3.156 | -4.4 | -4.4 | -4.4 | 2.694 | -4.4 | -1.029 | -4.4 | -0.97 | -4.4 | -3.147 | -4.4 | -1.332 | -4.4 | -4.4 | -4.4 |
| 14 | 1.015 | 1.711 | 1.29 | 1.267 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -1.618 | -3.134 | -1.768 | -4.4 | -4.4 | -4.4 | -4.4 |
| 15 | -0.747 | -0.746 | 0.557 | -2.269 | 0.271 | 1.062 | -0.143 | -0.37 | -1.013 | 0.191 | 0.552 | -0.947 | -1.223 | 0.323 | 0.268 | -0.271 |
| 16 | -1.485 | -0.481 | 0.354 | -1.761 | -0.946 | 1.144 | -1.036 | 0.73 | 0.175 | 0.614 | 0.625 | -0.29 | -1.932 | -0.391 | -0.303 | -0.955 |
| 17 | -0.512 | -0.45 | -0.501 | -2.223 | 0.625 | 0.038 | -1.743 | 1.163 | 0.019 | -0.029 | 0.245 | 0.113 | -1.19 | -0.125 | -0.109 | 0.259 |
| 18 | 0.332 | -0.867 | 0.517 | -1.202 | 0.151 | 0.248 | -0.864 | -0.41 | -0.096 | -0.202 | -0.104 | -1.113 | -0.807 | 0.73 | 0.939 | -0.438 |
| 19 | 0.187 | -0.502 | 0.518 | -0.863 | 0.596 | 0.049 | -1.335 | 0.383 | 0.281 | 0.298 | 0.82 | -0.489 | -1.366 | -0.695 | -0.014 | -1.617 |
| 20 | -0.188 | -0.033 | 0.933 | -1.339 | 0.233 | 0.161 | -0.734 | -0.535 | -0.116 | -0.017 | 0.811 | 0.073 | -0.793 | -0.602 | 0.272 | -0.949 |
| 21 | -0.173 | -0.093 | 0.321 | -1.193 | 0.293 | 0.356 | -1.948 | -0.271 | 0.258 | 0.64 | 0.761 | 0.224 | -1.124 | -0.316 | -0.295 | -0.671 |
| 22 | -0.27 | 0.2 | 0.33 | -0.857 | 0.329 | -0.118 | -1.771 | 0.923 | 0.094 | -0.263 | 0.384 | 0.069 | -1.108 | 0.035 | -0.507 | -0.15 |