Model info
| Transcription factor | Irf1 | ||||||||
| Model | IRF1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 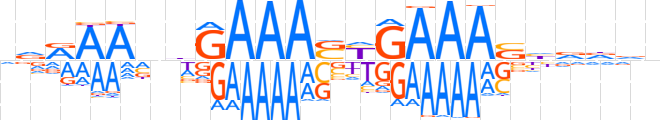 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnRAAnhGAAASYGAAAShvdvn | ||||||||
| Best auROC (human) | 0.997 | ||||||||
| Best auROC (mouse) | 0.998 | ||||||||
| Peak sets in benchmark (human) | 23 | ||||||||
| Peak sets in benchmark (mouse) | 18 | ||||||||
| Aligned words | 453 | ||||||||
| TF family | Interferon-regulatory factors {3.5.3} | ||||||||
| TF subfamily | IRF-1 {3.5.3.0.1} | ||||||||
| MGI | MGI:96590 | ||||||||
| EntrezGene | GeneID:16362 (SSTAR profile) | ||||||||
| UniProt ID | IRF1_MOUSE | ||||||||
| UniProt AC | P15314 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Irf1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 39.0 | 13.0 | 83.0 | 6.0 | 51.0 | 8.0 | 8.0 | 8.0 | 19.0 | 9.0 | 43.0 | 11.0 | 40.0 | 7.0 | 33.0 | 12.0 |
| 02 | 84.0 | 11.0 | 48.0 | 6.0 | 29.0 | 2.0 | 1.0 | 5.0 | 107.0 | 5.0 | 52.0 | 3.0 | 18.0 | 1.0 | 12.0 | 6.0 |
| 03 | 210.0 | 5.0 | 9.0 | 14.0 | 8.0 | 6.0 | 2.0 | 3.0 | 102.0 | 0.0 | 4.0 | 7.0 | 16.0 | 2.0 | 1.0 | 1.0 |
| 04 | 298.0 | 9.0 | 17.0 | 12.0 | 11.0 | 1.0 | 0.0 | 1.0 | 16.0 | 0.0 | 0.0 | 0.0 | 14.0 | 1.0 | 2.0 | 8.0 |
| 05 | 122.0 | 92.0 | 85.0 | 40.0 | 6.0 | 3.0 | 0.0 | 2.0 | 5.0 | 7.0 | 3.0 | 4.0 | 6.0 | 5.0 | 3.0 | 7.0 |
| 06 | 26.0 | 33.0 | 20.0 | 60.0 | 26.0 | 43.0 | 2.0 | 36.0 | 30.0 | 13.0 | 9.0 | 39.0 | 22.0 | 5.0 | 16.0 | 10.0 |
| 07 | 4.0 | 0.0 | 100.0 | 0.0 | 58.0 | 0.0 | 35.0 | 1.0 | 11.0 | 0.0 | 36.0 | 0.0 | 8.0 | 0.0 | 137.0 | 0.0 |
| 08 | 81.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 307.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 09 | 387.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 388.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 11 | 22.0 | 203.0 | 159.0 | 6.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 1.0 | 3.0 | 0.0 | 18.0 | 38.0 | 51.0 | 2.0 | 112.0 | 23.0 | 19.0 | 5.0 | 112.0 | 1.0 | 1.0 | 0.0 | 4.0 |
| 13 | 3.0 | 0.0 | 60.0 | 0.0 | 47.0 | 1.0 | 26.0 | 0.0 | 1.0 | 0.0 | 6.0 | 0.0 | 15.0 | 0.0 | 231.0 | 0.0 |
| 14 | 62.0 | 0.0 | 4.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 313.0 | 0.0 | 8.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 374.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 16 | 378.0 | 1.0 | 9.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| 17 | 18.0 | 157.0 | 189.0 | 15.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 4.0 | 5.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 18 | 1.0 | 5.0 | 1.0 | 11.0 | 16.0 | 74.0 | 5.0 | 67.0 | 33.0 | 42.0 | 25.0 | 94.0 | 4.0 | 7.0 | 4.0 | 1.0 |
| 19 | 24.0 | 6.0 | 24.0 | 0.0 | 88.0 | 10.0 | 14.0 | 16.0 | 16.0 | 4.0 | 13.0 | 2.0 | 55.0 | 24.0 | 74.0 | 20.0 |
| 20 | 107.0 | 17.0 | 36.0 | 23.0 | 22.0 | 6.0 | 7.0 | 9.0 | 56.0 | 15.0 | 36.0 | 18.0 | 15.0 | 7.0 | 3.0 | 13.0 |
| 21 | 107.0 | 30.0 | 53.0 | 10.0 | 19.0 | 11.0 | 3.0 | 12.0 | 25.0 | 13.0 | 20.0 | 24.0 | 19.0 | 12.0 | 26.0 | 6.0 |
| 22 | 79.0 | 42.0 | 32.0 | 17.0 | 17.0 | 19.0 | 9.0 | 21.0 | 31.0 | 31.0 | 23.0 | 17.0 | 15.0 | 10.0 | 10.0 | 17.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.464 | -0.616 | 1.215 | -1.357 | 0.73 | -1.084 | -1.084 | -1.084 | -0.245 | -0.971 | 0.561 | -0.778 | 0.489 | -1.211 | 0.299 | -0.693 |
| 02 | 1.227 | -0.778 | 0.67 | -1.357 | 0.171 | -2.345 | -2.892 | -1.527 | 1.468 | -1.527 | 0.75 | -1.993 | -0.298 | -2.892 | -0.693 | -1.357 |
| 03 | 2.14 | -1.527 | -0.971 | -0.543 | -1.084 | -1.357 | -2.345 | -1.993 | 1.42 | -4.195 | -1.733 | -1.211 | -0.413 | -2.345 | -2.892 | -2.892 |
| 04 | 2.49 | -0.971 | -0.354 | -0.693 | -0.778 | -2.892 | -4.195 | -2.892 | -0.413 | -4.195 | -4.195 | -4.195 | -0.543 | -2.892 | -2.345 | -1.084 |
| 05 | 1.598 | 1.317 | 1.238 | 0.489 | -1.357 | -1.993 | -4.195 | -2.345 | -1.527 | -1.211 | -1.993 | -1.733 | -1.357 | -1.527 | -1.993 | -1.211 |
| 06 | 0.064 | 0.299 | -0.195 | 0.892 | 0.064 | 0.561 | -2.345 | 0.385 | 0.205 | -0.616 | -0.971 | 0.464 | -0.101 | -1.527 | -0.413 | -0.87 |
| 07 | -1.733 | -4.195 | 1.4 | -4.195 | 0.858 | -4.195 | 0.357 | -2.892 | -0.778 | -4.195 | 0.385 | -4.195 | -1.084 | -4.195 | 1.714 | -4.195 |
| 08 | 1.19 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | 2.519 | -4.195 | -2.892 | -4.195 | -2.892 | -4.195 | -4.195 | -4.195 |
| 09 | 2.751 | -4.195 | -4.195 | -2.345 | -4.195 | -4.195 | -4.195 | -4.195 | -2.892 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 |
| 10 | 2.753 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -2.345 | -4.195 | -4.195 | -4.195 |
| 11 | -0.101 | 2.106 | 1.863 | -1.357 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 |
| 12 | -2.892 | -1.993 | -4.195 | -0.298 | 0.439 | 0.73 | -2.345 | 1.513 | -0.057 | -0.245 | -1.527 | 1.513 | -2.892 | -2.892 | -4.195 | -1.733 |
| 13 | -1.993 | -4.195 | 0.892 | -4.195 | 0.649 | -2.892 | 0.064 | -4.195 | -2.892 | -4.195 | -1.357 | -4.195 | -0.476 | -4.195 | 2.235 | -4.195 |
| 14 | 0.924 | -4.195 | -1.733 | -4.195 | -2.892 | -4.195 | -4.195 | -4.195 | 2.539 | -4.195 | -1.084 | -2.345 | -4.195 | -4.195 | -4.195 | -4.195 |
| 15 | 2.717 | -4.195 | -4.195 | -2.345 | -4.195 | -4.195 | -4.195 | -4.195 | -0.693 | -4.195 | -4.195 | -4.195 | -2.345 | -4.195 | -4.195 | -4.195 |
| 16 | 2.727 | -2.892 | -0.971 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -4.195 | -2.892 | -4.195 | -2.892 | -4.195 |
| 17 | -0.298 | 1.85 | 2.035 | -0.476 | -4.195 | -2.892 | -4.195 | -4.195 | -4.195 | -1.733 | -1.527 | -2.892 | -4.195 | -4.195 | -4.195 | -4.195 |
| 18 | -2.892 | -1.527 | -2.892 | -0.778 | -0.413 | 1.1 | -1.527 | 1.002 | 0.299 | 0.538 | 0.025 | 1.339 | -1.733 | -1.211 | -1.733 | -2.892 |
| 19 | -0.015 | -1.357 | -0.015 | -4.195 | 1.273 | -0.87 | -0.543 | -0.413 | -0.413 | -1.733 | -0.616 | -2.345 | 0.805 | -0.015 | 1.1 | -0.195 |
| 20 | 1.468 | -0.354 | 0.385 | -0.057 | -0.101 | -1.357 | -1.211 | -0.971 | 0.823 | -0.476 | 0.385 | -0.298 | -0.476 | -1.211 | -1.993 | -0.616 |
| 21 | 1.468 | 0.205 | 0.769 | -0.87 | -0.245 | -0.778 | -1.993 | -0.693 | 0.025 | -0.616 | -0.195 | -0.015 | -0.245 | -0.693 | 0.064 | -1.357 |
| 22 | 1.165 | 0.538 | 0.269 | -0.354 | -0.354 | -0.245 | -0.971 | -0.147 | 0.237 | 0.237 | -0.057 | -0.354 | -0.476 | -0.87 | -0.87 | -0.354 |