Model info
| Transcription factor | Prdm14 | ||||||||
| Model | PRD14_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 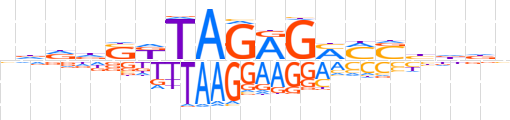 | ||||||||
| LOGO (reverse complement) | 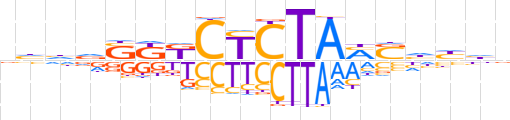 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndddSKTAGRGMMSbddn | ||||||||
| Best auROC (human) | 0.881 | ||||||||
| Best auROC (mouse) | 0.947 | ||||||||
| Peak sets in benchmark (human) | 7 | ||||||||
| Peak sets in benchmark (mouse) | 11 | ||||||||
| Aligned words | 535 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| MGI | MGI:3588194 | ||||||||
| EntrezGene | GeneID:383491 (SSTAR profile) | ||||||||
| UniProt ID | PRD14_MOUSE | ||||||||
| UniProt AC | E9Q3T6 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Prdm14 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 35.0 | 8.0 | 45.0 | 19.0 | 103.0 | 19.0 | 1.0 | 44.0 | 33.0 | 10.0 | 43.0 | 21.0 | 22.0 | 10.0 | 40.0 | 27.0 |
| 02 | 41.0 | 13.0 | 125.0 | 14.0 | 27.0 | 9.0 | 5.0 | 6.0 | 37.0 | 15.0 | 60.0 | 17.0 | 27.0 | 13.0 | 55.0 | 16.0 |
| 03 | 40.0 | 6.0 | 67.0 | 19.0 | 29.0 | 6.0 | 3.0 | 12.0 | 84.0 | 11.0 | 64.0 | 86.0 | 10.0 | 1.0 | 34.0 | 8.0 |
| 04 | 10.0 | 35.0 | 106.0 | 12.0 | 6.0 | 5.0 | 7.0 | 6.0 | 11.0 | 34.0 | 103.0 | 20.0 | 6.0 | 15.0 | 103.0 | 1.0 |
| 05 | 8.0 | 0.0 | 8.0 | 17.0 | 15.0 | 2.0 | 5.0 | 67.0 | 58.0 | 6.0 | 89.0 | 166.0 | 6.0 | 1.0 | 3.0 | 29.0 |
| 06 | 9.0 | 7.0 | 2.0 | 69.0 | 2.0 | 0.0 | 0.0 | 7.0 | 6.0 | 2.0 | 3.0 | 94.0 | 1.0 | 0.0 | 8.0 | 270.0 |
| 07 | 18.0 | 0.0 | 0.0 | 0.0 | 9.0 | 0.0 | 0.0 | 0.0 | 11.0 | 0.0 | 0.0 | 2.0 | 424.0 | 4.0 | 5.0 | 7.0 |
| 08 | 32.0 | 22.0 | 408.0 | 0.0 | 3.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 4.0 | 0.0 | 2.0 | 1.0 | 6.0 | 0.0 |
| 09 | 30.0 | 0.0 | 7.0 | 1.0 | 21.0 | 0.0 | 1.0 | 1.0 | 311.0 | 7.0 | 85.0 | 16.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 10.0 | 11.0 | 328.0 | 13.0 | 0.0 | 0.0 | 6.0 | 1.0 | 6.0 | 2.0 | 84.0 | 1.0 | 0.0 | 0.0 | 18.0 | 0.0 |
| 11 | 7.0 | 6.0 | 1.0 | 2.0 | 8.0 | 1.0 | 0.0 | 4.0 | 266.0 | 121.0 | 22.0 | 27.0 | 10.0 | 2.0 | 1.0 | 2.0 |
| 12 | 46.0 | 202.0 | 17.0 | 26.0 | 32.0 | 82.0 | 0.0 | 16.0 | 5.0 | 12.0 | 3.0 | 4.0 | 15.0 | 17.0 | 1.0 | 2.0 |
| 13 | 12.0 | 50.0 | 25.0 | 11.0 | 30.0 | 247.0 | 16.0 | 20.0 | 0.0 | 14.0 | 3.0 | 4.0 | 3.0 | 18.0 | 26.0 | 1.0 |
| 14 | 9.0 | 17.0 | 12.0 | 7.0 | 35.0 | 161.0 | 12.0 | 121.0 | 7.0 | 21.0 | 25.0 | 17.0 | 3.0 | 13.0 | 8.0 | 12.0 |
| 15 | 10.0 | 4.0 | 24.0 | 16.0 | 62.0 | 17.0 | 8.0 | 125.0 | 20.0 | 4.0 | 17.0 | 16.0 | 27.0 | 13.0 | 70.0 | 47.0 |
| 16 | 16.0 | 21.0 | 62.0 | 20.0 | 9.0 | 11.0 | 4.0 | 14.0 | 34.0 | 20.0 | 42.0 | 23.0 | 22.0 | 19.0 | 133.0 | 30.0 |
| 17 | 26.0 | 13.0 | 30.0 | 12.0 | 23.0 | 26.0 | 5.0 | 17.0 | 27.0 | 112.0 | 62.0 | 40.0 | 17.0 | 28.0 | 18.0 | 24.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.152 | -1.287 | 0.401 | -0.449 | 1.224 | -0.449 | -3.088 | 0.379 | 0.094 | -1.074 | 0.356 | -0.351 | -0.306 | -1.074 | 0.285 | -0.104 |
| 02 | 0.309 | -0.82 | 1.417 | -0.748 | -0.104 | -1.175 | -1.73 | -1.56 | 0.207 | -0.681 | 0.687 | -0.558 | -0.104 | -0.82 | 0.6 | -0.618 |
| 03 | 0.285 | -1.56 | 0.796 | -0.449 | -0.033 | -1.56 | -2.194 | -0.897 | 1.021 | -0.982 | 0.751 | 1.045 | -1.074 | -3.088 | 0.124 | -1.287 |
| 04 | -1.074 | 0.152 | 1.253 | -0.897 | -1.56 | -1.73 | -1.414 | -1.56 | -0.982 | 0.124 | 1.224 | -0.399 | -1.56 | -0.681 | 1.224 | -3.088 |
| 05 | -1.287 | -4.366 | -1.287 | -0.558 | -0.681 | -2.544 | -1.73 | 0.796 | 0.653 | -1.56 | 1.079 | 1.7 | -1.56 | -3.088 | -2.194 | -0.033 |
| 06 | -1.175 | -1.414 | -2.544 | 0.826 | -2.544 | -4.366 | -4.366 | -1.414 | -1.56 | -2.544 | -2.194 | 1.133 | -3.088 | -4.366 | -1.287 | 2.186 |
| 07 | -0.502 | -4.366 | -4.366 | -4.366 | -1.175 | -4.366 | -4.366 | -4.366 | -0.982 | -4.366 | -4.366 | -2.544 | 2.637 | -1.936 | -1.73 | -1.414 |
| 08 | 0.064 | -0.306 | 2.598 | -4.366 | -2.194 | -4.366 | -3.088 | -4.366 | -3.088 | -4.366 | -1.936 | -4.366 | -2.544 | -3.088 | -1.56 | -4.366 |
| 09 | 0.0 | -4.366 | -1.414 | -3.088 | -0.351 | -4.366 | -3.088 | -3.088 | 2.327 | -1.414 | 1.033 | -0.618 | -4.366 | -4.366 | -4.366 | -4.366 |
| 10 | -1.074 | -0.982 | 2.38 | -0.82 | -4.366 | -4.366 | -1.56 | -3.088 | -1.56 | -2.544 | 1.021 | -3.088 | -4.366 | -4.366 | -0.502 | -4.366 |
| 11 | -1.414 | -1.56 | -3.088 | -2.544 | -1.287 | -3.088 | -4.366 | -1.936 | 2.171 | 1.385 | -0.306 | -0.104 | -1.074 | -2.544 | -3.088 | -2.544 |
| 12 | 0.423 | 1.896 | -0.558 | -0.141 | 0.064 | 0.997 | -4.366 | -0.618 | -1.73 | -0.897 | -2.194 | -1.936 | -0.681 | -0.558 | -3.088 | -2.544 |
| 13 | -0.897 | 0.506 | -0.18 | -0.982 | 0.0 | 2.097 | -0.618 | -0.399 | -4.366 | -0.748 | -2.194 | -1.936 | -2.194 | -0.502 | -0.141 | -3.088 |
| 14 | -1.175 | -0.558 | -0.897 | -1.414 | 0.152 | 1.67 | -0.897 | 1.385 | -1.414 | -0.351 | -0.18 | -0.558 | -2.194 | -0.82 | -1.287 | -0.897 |
| 15 | -1.074 | -1.936 | -0.22 | -0.618 | 0.719 | -0.558 | -1.287 | 1.417 | -0.399 | -1.936 | -0.558 | -0.618 | -0.104 | -0.82 | 0.84 | 0.444 |
| 16 | -0.618 | -0.351 | 0.719 | -0.399 | -1.175 | -0.982 | -1.936 | -0.748 | 0.124 | -0.399 | 0.333 | -0.262 | -0.306 | -0.449 | 1.479 | 0.0 |
| 17 | -0.141 | -0.82 | 0.0 | -0.897 | -0.262 | -0.141 | -1.73 | -0.558 | -0.104 | 1.308 | 0.719 | 0.285 | -0.558 | -0.068 | -0.502 | -0.22 |