Model info
| Transcription factor | Rxrg | ||||||||
| Model | RXRG_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 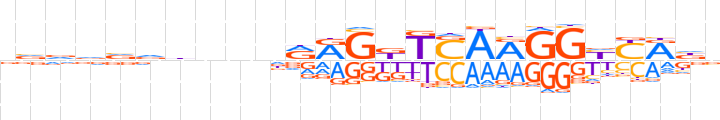 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 25 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dvddvdbnnnRRGKKSARGGYYRnv | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.986 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 7 | ||||||||
| Aligned words | 395 | ||||||||
| TF family | RXR-related receptors (NR2) {2.1.3} | ||||||||
| TF subfamily | Retinoid X receptors (NR2B) {2.1.3.1} | ||||||||
| MGI | MGI:98216 | ||||||||
| EntrezGene | GeneID:20183 (SSTAR profile) | ||||||||
| UniProt ID | RXRG_MOUSE | ||||||||
| UniProt AC | P28705 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Rxrg expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 13.0 | 13.0 | 48.0 | 9.0 | 13.0 | 14.0 | 6.0 | 18.0 | 37.0 | 30.0 | 107.0 | 19.0 | 10.0 | 9.0 | 32.0 | 10.0 |
| 02 | 30.0 | 9.0 | 29.0 | 5.0 | 31.0 | 7.0 | 4.0 | 24.0 | 77.0 | 14.0 | 71.0 | 31.0 | 5.0 | 5.0 | 36.0 | 10.0 |
| 03 | 44.0 | 12.0 | 71.0 | 16.0 | 14.0 | 4.0 | 2.0 | 15.0 | 38.0 | 9.0 | 63.0 | 30.0 | 16.0 | 23.0 | 16.0 | 15.0 |
| 04 | 11.0 | 14.0 | 77.0 | 10.0 | 19.0 | 9.0 | 9.0 | 11.0 | 33.0 | 11.0 | 88.0 | 20.0 | 10.0 | 29.0 | 25.0 | 12.0 |
| 05 | 27.0 | 11.0 | 23.0 | 12.0 | 33.0 | 10.0 | 8.0 | 12.0 | 65.0 | 15.0 | 90.0 | 29.0 | 6.0 | 10.0 | 28.0 | 9.0 |
| 06 | 17.0 | 30.0 | 36.0 | 48.0 | 11.0 | 11.0 | 0.0 | 24.0 | 13.0 | 26.0 | 50.0 | 60.0 | 5.0 | 27.0 | 14.0 | 16.0 |
| 07 | 9.0 | 5.0 | 20.0 | 12.0 | 30.0 | 23.0 | 7.0 | 34.0 | 29.0 | 23.0 | 25.0 | 23.0 | 21.0 | 74.0 | 37.0 | 16.0 |
| 08 | 17.0 | 15.0 | 45.0 | 12.0 | 73.0 | 33.0 | 9.0 | 10.0 | 19.0 | 12.0 | 37.0 | 21.0 | 5.0 | 37.0 | 30.0 | 13.0 |
| 09 | 21.0 | 12.0 | 67.0 | 14.0 | 44.0 | 12.0 | 6.0 | 35.0 | 54.0 | 12.0 | 21.0 | 34.0 | 6.0 | 23.0 | 16.0 | 11.0 |
| 10 | 36.0 | 4.0 | 73.0 | 12.0 | 45.0 | 4.0 | 6.0 | 4.0 | 38.0 | 4.0 | 65.0 | 3.0 | 16.0 | 7.0 | 64.0 | 7.0 |
| 11 | 98.0 | 2.0 | 29.0 | 6.0 | 12.0 | 2.0 | 3.0 | 2.0 | 130.0 | 5.0 | 69.0 | 4.0 | 8.0 | 1.0 | 11.0 | 6.0 |
| 12 | 9.0 | 12.0 | 222.0 | 5.0 | 6.0 | 1.0 | 2.0 | 1.0 | 11.0 | 18.0 | 82.0 | 1.0 | 2.0 | 1.0 | 13.0 | 2.0 |
| 13 | 2.0 | 8.0 | 14.0 | 4.0 | 17.0 | 2.0 | 5.0 | 8.0 | 8.0 | 15.0 | 137.0 | 159.0 | 0.0 | 3.0 | 4.0 | 2.0 |
| 14 | 1.0 | 5.0 | 19.0 | 2.0 | 2.0 | 1.0 | 3.0 | 22.0 | 3.0 | 11.0 | 24.0 | 122.0 | 3.0 | 0.0 | 14.0 | 156.0 |
| 15 | 1.0 | 5.0 | 3.0 | 0.0 | 2.0 | 13.0 | 1.0 | 1.0 | 7.0 | 21.0 | 24.0 | 8.0 | 14.0 | 251.0 | 28.0 | 9.0 |
| 16 | 14.0 | 2.0 | 5.0 | 3.0 | 269.0 | 4.0 | 6.0 | 11.0 | 36.0 | 5.0 | 8.0 | 7.0 | 7.0 | 5.0 | 4.0 | 2.0 |
| 17 | 238.0 | 20.0 | 51.0 | 17.0 | 15.0 | 0.0 | 0.0 | 1.0 | 19.0 | 2.0 | 1.0 | 1.0 | 10.0 | 4.0 | 6.0 | 3.0 |
| 18 | 17.0 | 3.0 | 259.0 | 3.0 | 7.0 | 0.0 | 14.0 | 5.0 | 7.0 | 6.0 | 41.0 | 4.0 | 5.0 | 5.0 | 8.0 | 4.0 |
| 19 | 0.0 | 5.0 | 28.0 | 3.0 | 0.0 | 8.0 | 4.0 | 2.0 | 14.0 | 9.0 | 290.0 | 9.0 | 0.0 | 4.0 | 10.0 | 2.0 |
| 20 | 0.0 | 4.0 | 3.0 | 7.0 | 4.0 | 2.0 | 0.0 | 20.0 | 37.0 | 68.0 | 29.0 | 198.0 | 1.0 | 2.0 | 7.0 | 6.0 |
| 21 | 1.0 | 23.0 | 15.0 | 3.0 | 4.0 | 56.0 | 1.0 | 15.0 | 5.0 | 21.0 | 10.0 | 3.0 | 5.0 | 176.0 | 22.0 | 28.0 |
| 22 | 6.0 | 0.0 | 3.0 | 6.0 | 234.0 | 20.0 | 10.0 | 12.0 | 18.0 | 5.0 | 19.0 | 6.0 | 23.0 | 1.0 | 19.0 | 6.0 |
| 23 | 35.0 | 37.0 | 153.0 | 56.0 | 8.0 | 2.0 | 4.0 | 12.0 | 5.0 | 3.0 | 30.0 | 13.0 | 2.0 | 8.0 | 17.0 | 3.0 |
| 24 | 11.0 | 12.0 | 24.0 | 3.0 | 10.0 | 29.0 | 0.0 | 11.0 | 34.0 | 68.0 | 81.0 | 21.0 | 4.0 | 40.0 | 23.0 | 17.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.61 | -0.61 | 0.675 | -0.966 | -0.61 | -0.538 | -1.352 | -0.293 | 0.417 | 0.21 | 1.473 | -0.24 | -0.864 | -0.966 | 0.274 | -0.864 |
| 02 | 0.21 | -0.966 | 0.176 | -1.522 | 0.242 | -1.206 | -1.728 | -0.01 | 1.145 | -0.538 | 1.064 | 0.242 | -1.522 | -1.522 | 0.39 | -0.864 |
| 03 | 0.589 | -0.688 | 1.064 | -0.408 | -0.538 | -1.728 | -2.34 | -0.471 | 0.444 | -0.966 | 0.945 | 0.21 | -0.408 | -0.052 | -0.408 | -0.471 |
| 04 | -0.772 | -0.538 | 1.145 | -0.864 | -0.24 | -0.966 | -0.966 | -0.772 | 0.304 | -0.772 | 1.278 | -0.189 | -0.864 | 0.176 | 0.03 | -0.688 |
| 05 | 0.106 | -0.772 | -0.052 | -0.688 | 0.304 | -0.864 | -1.079 | -0.688 | 0.976 | -0.471 | 1.3 | 0.176 | -1.352 | -0.864 | 0.142 | -0.966 |
| 06 | -0.349 | 0.21 | 0.39 | 0.675 | -0.772 | -0.772 | -4.191 | -0.01 | -0.61 | 0.069 | 0.716 | 0.897 | -1.522 | 0.106 | -0.538 | -0.408 |
| 07 | -0.966 | -1.522 | -0.189 | -0.688 | 0.21 | -0.052 | -1.206 | 0.334 | 0.176 | -0.052 | 0.03 | -0.052 | -0.142 | 1.105 | 0.417 | -0.408 |
| 08 | -0.349 | -0.471 | 0.611 | -0.688 | 1.092 | 0.304 | -0.966 | -0.864 | -0.24 | -0.688 | 0.417 | -0.142 | -1.522 | 0.417 | 0.21 | -0.61 |
| 09 | -0.142 | -0.688 | 1.007 | -0.538 | 0.589 | -0.688 | -1.352 | 0.362 | 0.792 | -0.688 | -0.142 | 0.334 | -1.352 | -0.052 | -0.408 | -0.772 |
| 10 | 0.39 | -1.728 | 1.092 | -0.688 | 0.611 | -1.728 | -1.352 | -1.728 | 0.444 | -1.728 | 0.976 | -1.988 | -0.408 | -1.206 | 0.961 | -1.206 |
| 11 | 1.385 | -2.34 | 0.176 | -1.352 | -0.688 | -2.34 | -1.988 | -2.34 | 1.667 | -1.522 | 1.036 | -1.728 | -1.079 | -2.887 | -0.772 | -1.352 |
| 12 | -0.966 | -0.688 | 2.201 | -1.522 | -1.352 | -2.887 | -2.34 | -2.887 | -0.772 | -0.293 | 1.208 | -2.887 | -2.34 | -2.887 | -0.61 | -2.34 |
| 13 | -2.34 | -1.079 | -0.538 | -1.728 | -0.349 | -2.34 | -1.522 | -1.079 | -1.079 | -0.471 | 1.719 | 1.868 | -4.191 | -1.988 | -1.728 | -2.34 |
| 14 | -2.887 | -1.522 | -0.24 | -2.34 | -2.34 | -2.887 | -1.988 | -0.096 | -1.988 | -0.772 | -0.01 | 1.603 | -1.988 | -4.191 | -0.538 | 1.849 |
| 15 | -2.887 | -1.522 | -1.988 | -4.191 | -2.34 | -0.61 | -2.887 | -2.887 | -1.206 | -0.142 | -0.01 | -1.079 | -0.538 | 2.323 | 0.142 | -0.966 |
| 16 | -0.538 | -2.34 | -1.522 | -1.988 | 2.392 | -1.728 | -1.352 | -0.772 | 0.39 | -1.522 | -1.079 | -1.206 | -1.206 | -1.522 | -1.728 | -2.34 |
| 17 | 2.27 | -0.189 | 0.735 | -0.349 | -0.471 | -4.191 | -4.191 | -2.887 | -0.24 | -2.34 | -2.887 | -2.887 | -0.864 | -1.728 | -1.352 | -1.988 |
| 18 | -0.349 | -1.988 | 2.355 | -1.988 | -1.206 | -4.191 | -0.538 | -1.522 | -1.206 | -1.352 | 0.519 | -1.728 | -1.522 | -1.522 | -1.079 | -1.728 |
| 19 | -4.191 | -1.522 | 0.142 | -1.988 | -4.191 | -1.079 | -1.728 | -2.34 | -0.538 | -0.966 | 2.468 | -0.966 | -4.191 | -1.728 | -0.864 | -2.34 |
| 20 | -4.191 | -1.728 | -1.988 | -1.206 | -1.728 | -2.34 | -4.191 | -0.189 | 0.417 | 1.021 | 0.176 | 2.086 | -2.887 | -2.34 | -1.206 | -1.352 |
| 21 | -2.887 | -0.052 | -0.471 | -1.988 | -1.728 | 0.828 | -2.887 | -0.471 | -1.522 | -0.142 | -0.864 | -1.988 | -1.522 | 1.969 | -0.096 | 0.142 |
| 22 | -1.352 | -4.191 | -1.988 | -1.352 | 2.253 | -0.189 | -0.864 | -0.688 | -0.293 | -1.522 | -0.24 | -1.352 | -0.052 | -2.887 | -0.24 | -1.352 |
| 23 | 0.362 | 0.417 | 1.829 | 0.828 | -1.079 | -2.34 | -1.728 | -0.688 | -1.522 | -1.988 | 0.21 | -0.61 | -2.34 | -1.079 | -0.349 | -1.988 |
| 24 | -0.772 | -0.688 | -0.01 | -1.988 | -0.864 | 0.176 | -4.191 | -0.772 | 0.334 | 1.021 | 1.195 | -0.142 | -1.728 | 0.494 | -0.052 | -0.349 |