Model info
| Transcription factor | Sp1 | ||||||||
| Model | SP1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 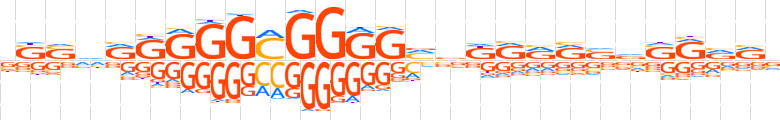 | ||||||||
| LOGO (reverse complement) | 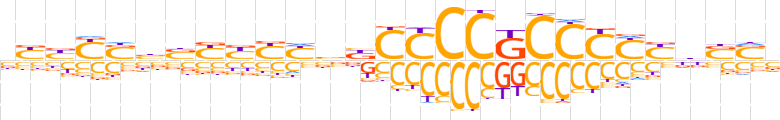 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 27 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vSSvRRGGGMGGGGvvdRRRSvSRvRv | ||||||||
| Best auROC (human) | 0.999 | ||||||||
| Best auROC (mouse) | 0.991 | ||||||||
| Peak sets in benchmark (human) | 53 | ||||||||
| Peak sets in benchmark (mouse) | 11 | ||||||||
| Aligned words | 494 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | Sp1-like factors {2.3.1.1} | ||||||||
| MGI | MGI:98372 | ||||||||
| EntrezGene | GeneID:20683 (SSTAR profile) | ||||||||
| UniProt ID | SP1_MOUSE | ||||||||
| UniProt AC | O89090 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Sp1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 5.0 | 11.0 | 45.0 | 1.0 | 7.0 | 12.0 | 75.0 | 16.0 | 21.0 | 44.0 | 176.0 | 26.0 | 6.0 | 2.0 | 36.0 | 10.0 |
| 02 | 8.0 | 14.0 | 15.0 | 2.0 | 13.0 | 11.0 | 42.0 | 3.0 | 31.0 | 62.0 | 225.0 | 14.0 | 5.0 | 13.0 | 30.0 | 5.0 |
| 03 | 29.0 | 6.0 | 17.0 | 5.0 | 38.0 | 19.0 | 10.0 | 33.0 | 134.0 | 75.0 | 64.0 | 39.0 | 2.0 | 9.0 | 3.0 | 10.0 |
| 04 | 32.0 | 21.0 | 147.0 | 3.0 | 16.0 | 13.0 | 77.0 | 3.0 | 27.0 | 19.0 | 41.0 | 7.0 | 18.0 | 12.0 | 55.0 | 2.0 |
| 05 | 11.0 | 2.0 | 75.0 | 5.0 | 2.0 | 8.0 | 48.0 | 7.0 | 31.0 | 23.0 | 237.0 | 29.0 | 2.0 | 0.0 | 12.0 | 1.0 |
| 06 | 10.0 | 4.0 | 31.0 | 1.0 | 2.0 | 2.0 | 28.0 | 1.0 | 53.0 | 9.0 | 300.0 | 10.0 | 6.0 | 2.0 | 34.0 | 0.0 |
| 07 | 8.0 | 2.0 | 59.0 | 2.0 | 2.0 | 2.0 | 9.0 | 4.0 | 13.0 | 6.0 | 362.0 | 12.0 | 3.0 | 3.0 | 5.0 | 1.0 |
| 08 | 4.0 | 1.0 | 17.0 | 4.0 | 1.0 | 0.0 | 12.0 | 0.0 | 6.0 | 7.0 | 416.0 | 6.0 | 1.0 | 3.0 | 13.0 | 2.0 |
| 09 | 5.0 | 7.0 | 0.0 | 0.0 | 1.0 | 8.0 | 0.0 | 2.0 | 131.0 | 318.0 | 2.0 | 7.0 | 3.0 | 9.0 | 0.0 | 0.0 |
| 10 | 13.0 | 0.0 | 127.0 | 0.0 | 12.0 | 0.0 | 325.0 | 5.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 9.0 | 0.0 |
| 11 | 1.0 | 0.0 | 22.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 452.0 | 3.0 | 1.0 | 0.0 | 4.0 | 0.0 |
| 12 | 2.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 69.0 | 7.0 | 385.0 | 17.0 | 0.0 | 1.0 | 4.0 | 1.0 |
| 13 | 9.0 | 3.0 | 55.0 | 4.0 | 1.0 | 2.0 | 5.0 | 0.0 | 35.0 | 11.0 | 340.0 | 10.0 | 4.0 | 0.0 | 14.0 | 0.0 |
| 14 | 7.0 | 22.0 | 16.0 | 4.0 | 3.0 | 5.0 | 6.0 | 2.0 | 109.0 | 236.0 | 47.0 | 22.0 | 1.0 | 8.0 | 2.0 | 3.0 |
| 15 | 26.0 | 14.0 | 72.0 | 8.0 | 29.0 | 106.0 | 92.0 | 44.0 | 25.0 | 19.0 | 14.0 | 13.0 | 5.0 | 14.0 | 7.0 | 5.0 |
| 16 | 15.0 | 4.0 | 57.0 | 9.0 | 14.0 | 9.0 | 105.0 | 25.0 | 34.0 | 14.0 | 118.0 | 19.0 | 5.0 | 4.0 | 46.0 | 15.0 |
| 17 | 17.0 | 7.0 | 43.0 | 1.0 | 6.0 | 3.0 | 17.0 | 5.0 | 48.0 | 39.0 | 225.0 | 14.0 | 10.0 | 2.0 | 52.0 | 4.0 |
| 18 | 19.0 | 8.0 | 47.0 | 7.0 | 5.0 | 10.0 | 34.0 | 2.0 | 100.0 | 46.0 | 180.0 | 11.0 | 4.0 | 2.0 | 17.0 | 1.0 |
| 19 | 22.0 | 15.0 | 89.0 | 2.0 | 13.0 | 7.0 | 42.0 | 4.0 | 41.0 | 49.0 | 178.0 | 10.0 | 1.0 | 5.0 | 12.0 | 3.0 |
| 20 | 18.0 | 15.0 | 39.0 | 5.0 | 0.0 | 16.0 | 51.0 | 9.0 | 34.0 | 94.0 | 177.0 | 16.0 | 0.0 | 5.0 | 13.0 | 1.0 |
| 21 | 14.0 | 5.0 | 25.0 | 8.0 | 16.0 | 23.0 | 73.0 | 18.0 | 57.0 | 76.0 | 126.0 | 21.0 | 3.0 | 6.0 | 16.0 | 6.0 |
| 22 | 11.0 | 7.0 | 64.0 | 8.0 | 12.0 | 11.0 | 74.0 | 13.0 | 33.0 | 44.0 | 152.0 | 11.0 | 6.0 | 2.0 | 41.0 | 4.0 |
| 23 | 13.0 | 3.0 | 40.0 | 6.0 | 4.0 | 3.0 | 47.0 | 10.0 | 34.0 | 35.0 | 256.0 | 6.0 | 5.0 | 5.0 | 22.0 | 4.0 |
| 24 | 16.0 | 11.0 | 28.0 | 1.0 | 4.0 | 5.0 | 31.0 | 6.0 | 99.0 | 53.0 | 191.0 | 22.0 | 5.0 | 5.0 | 15.0 | 1.0 |
| 25 | 27.0 | 13.0 | 80.0 | 4.0 | 9.0 | 13.0 | 43.0 | 9.0 | 53.0 | 45.0 | 158.0 | 9.0 | 2.0 | 4.0 | 21.0 | 3.0 |
| 26 | 14.0 | 23.0 | 47.0 | 7.0 | 7.0 | 16.0 | 40.0 | 12.0 | 53.0 | 86.0 | 146.0 | 17.0 | 0.0 | 9.0 | 6.0 | 10.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.756 | -1.008 | 0.375 | -3.113 | -1.441 | -0.924 | 0.882 | -0.644 | -0.378 | 0.353 | 1.732 | -0.168 | -1.586 | -2.57 | 0.154 | -1.1 |
| 02 | -1.314 | -0.774 | -0.707 | -2.57 | -0.846 | -1.008 | 0.306 | -2.22 | 0.006 | 0.693 | 1.977 | -0.774 | -1.756 | -0.846 | -0.026 | -1.756 |
| 03 | -0.06 | -1.586 | -0.585 | -1.756 | 0.207 | -0.476 | -1.1 | 0.068 | 1.46 | 0.882 | 0.725 | 0.233 | -2.57 | -1.201 | -2.22 | -1.1 |
| 04 | 0.037 | -0.378 | 1.553 | -2.22 | -0.644 | -0.846 | 0.908 | -2.22 | -0.13 | -0.476 | 0.283 | -1.441 | -0.529 | -0.924 | 0.574 | -2.57 |
| 05 | -1.008 | -2.57 | 0.882 | -1.756 | -2.57 | -1.314 | 0.439 | -1.441 | 0.006 | -0.288 | 2.029 | -0.06 | -2.57 | -4.388 | -0.924 | -3.113 |
| 06 | -1.1 | -1.962 | 0.006 | -3.113 | -2.57 | -2.57 | -0.094 | -3.113 | 0.537 | -1.201 | 2.265 | -1.1 | -1.586 | -2.57 | 0.097 | -4.388 |
| 07 | -1.314 | -2.57 | 0.644 | -2.57 | -2.57 | -2.57 | -1.201 | -1.962 | -0.846 | -1.586 | 2.452 | -0.924 | -2.22 | -2.22 | -1.756 | -3.113 |
| 08 | -1.962 | -3.113 | -0.585 | -1.962 | -3.113 | -4.388 | -0.924 | -4.388 | -1.586 | -1.441 | 2.591 | -1.586 | -3.113 | -2.22 | -0.846 | -2.57 |
| 09 | -1.756 | -1.441 | -4.388 | -4.388 | -3.113 | -1.314 | -4.388 | -2.57 | 1.438 | 2.323 | -2.57 | -1.441 | -2.22 | -1.201 | -4.388 | -4.388 |
| 10 | -0.846 | -4.388 | 1.407 | -4.388 | -0.924 | -4.388 | 2.345 | -1.756 | -3.113 | -4.388 | -3.113 | -4.388 | -4.388 | -4.388 | -1.201 | -4.388 |
| 11 | -3.113 | -4.388 | -0.332 | -2.22 | -4.388 | -4.388 | -4.388 | -4.388 | -1.441 | -4.388 | 2.674 | -2.22 | -3.113 | -4.388 | -1.962 | -4.388 |
| 12 | -2.57 | -4.388 | -1.441 | -4.388 | -4.388 | -4.388 | -4.388 | -4.388 | 0.799 | -1.441 | 2.514 | -0.585 | -4.388 | -3.113 | -1.962 | -3.113 |
| 13 | -1.201 | -2.22 | 0.574 | -1.962 | -3.113 | -2.57 | -1.756 | -4.388 | 0.126 | -1.008 | 2.39 | -1.1 | -1.962 | -4.388 | -0.774 | -4.388 |
| 14 | -1.441 | -0.332 | -0.644 | -1.962 | -2.22 | -1.756 | -1.586 | -2.57 | 1.254 | 2.025 | 0.418 | -0.332 | -3.113 | -1.314 | -2.57 | -2.22 |
| 15 | -0.168 | -0.774 | 0.842 | -1.314 | -0.06 | 1.227 | 1.086 | 0.353 | -0.206 | -0.476 | -0.774 | -0.846 | -1.756 | -0.774 | -1.441 | -1.756 |
| 16 | -0.707 | -1.962 | 0.609 | -1.201 | -0.774 | -1.201 | 1.217 | -0.206 | 0.097 | -0.774 | 1.334 | -0.476 | -1.756 | -1.962 | 0.397 | -0.707 |
| 17 | -0.585 | -1.441 | 0.33 | -3.113 | -1.586 | -2.22 | -0.585 | -1.756 | 0.439 | 0.233 | 1.977 | -0.774 | -1.1 | -2.57 | 0.518 | -1.962 |
| 18 | -0.476 | -1.314 | 0.418 | -1.441 | -1.756 | -1.1 | 0.097 | -2.57 | 1.169 | 0.397 | 1.755 | -1.008 | -1.962 | -2.57 | -0.585 | -3.113 |
| 19 | -0.332 | -0.707 | 1.053 | -2.57 | -0.846 | -1.441 | 0.306 | -1.962 | 0.283 | 0.459 | 1.744 | -1.1 | -3.113 | -1.756 | -0.924 | -2.22 |
| 20 | -0.529 | -0.707 | 0.233 | -1.756 | -4.388 | -0.644 | 0.499 | -1.201 | 0.097 | 1.107 | 1.738 | -0.644 | -4.388 | -1.756 | -0.846 | -3.113 |
| 21 | -0.774 | -1.756 | -0.206 | -1.314 | -0.644 | -0.288 | 0.855 | -0.529 | 0.609 | 0.895 | 1.399 | -0.378 | -2.22 | -1.586 | -0.644 | -1.586 |
| 22 | -1.008 | -1.441 | 0.725 | -1.314 | -0.924 | -1.008 | 0.869 | -0.846 | 0.068 | 0.353 | 1.586 | -1.008 | -1.586 | -2.57 | 0.283 | -1.962 |
| 23 | -0.846 | -2.22 | 0.258 | -1.586 | -1.962 | -2.22 | 0.418 | -1.1 | 0.097 | 0.126 | 2.106 | -1.586 | -1.756 | -1.756 | -0.332 | -1.962 |
| 24 | -0.644 | -1.008 | -0.094 | -3.113 | -1.962 | -1.756 | 0.006 | -1.586 | 1.159 | 0.537 | 1.814 | -0.332 | -1.756 | -1.756 | -0.707 | -3.113 |
| 25 | -0.13 | -0.846 | 0.946 | -1.962 | -1.201 | -0.846 | 0.33 | -1.201 | 0.537 | 0.375 | 1.625 | -1.201 | -2.57 | -1.962 | -0.378 | -2.22 |
| 26 | -0.774 | -0.288 | 0.418 | -1.441 | -1.441 | -0.644 | 0.258 | -0.924 | 0.537 | 1.018 | 1.546 | -0.585 | -4.388 | -1.201 | -1.586 | -1.1 |