Model info
| Transcription factor | Tbx20 | ||||||||
| Model | TBX20_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 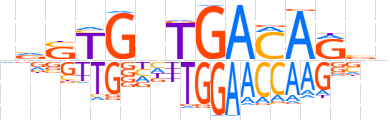 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvSTGnTGACARvn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.91 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 4 | ||||||||
| Aligned words | 468 | ||||||||
| TF family | TBX1-related factors {6.5.3} | ||||||||
| TF subfamily | TBX20 {6.5.3.0.5} | ||||||||
| MGI | MGI:1888496 | ||||||||
| EntrezGene | GeneID:57246 (SSTAR profile) | ||||||||
| UniProt ID | TBX20_MOUSE | ||||||||
| UniProt AC | Q9ES03 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Tbx20 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 37.0 | 13.0 | 98.0 | 15.0 | 26.0 | 15.0 | 12.0 | 26.0 | 41.0 | 17.0 | 61.0 | 5.0 | 9.0 | 16.0 | 66.0 | 9.0 |
| 02 | 6.0 | 14.0 | 93.0 | 0.0 | 8.0 | 26.0 | 16.0 | 11.0 | 2.0 | 73.0 | 145.0 | 17.0 | 2.0 | 12.0 | 37.0 | 4.0 |
| 03 | 1.0 | 1.0 | 0.0 | 16.0 | 18.0 | 15.0 | 0.0 | 92.0 | 24.0 | 24.0 | 2.0 | 241.0 | 0.0 | 5.0 | 0.0 | 27.0 |
| 04 | 6.0 | 2.0 | 35.0 | 0.0 | 3.0 | 0.0 | 39.0 | 3.0 | 0.0 | 0.0 | 2.0 | 0.0 | 7.0 | 3.0 | 364.0 | 2.0 |
| 05 | 5.0 | 3.0 | 5.0 | 3.0 | 2.0 | 3.0 | 0.0 | 0.0 | 107.0 | 123.0 | 87.0 | 123.0 | 1.0 | 1.0 | 2.0 | 1.0 |
| 06 | 7.0 | 3.0 | 1.0 | 104.0 | 4.0 | 2.0 | 1.0 | 123.0 | 1.0 | 4.0 | 3.0 | 86.0 | 4.0 | 1.0 | 20.0 | 102.0 |
| 07 | 0.0 | 0.0 | 16.0 | 0.0 | 1.0 | 0.0 | 9.0 | 0.0 | 3.0 | 0.0 | 22.0 | 0.0 | 7.0 | 1.0 | 407.0 | 0.0 |
| 08 | 10.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 445.0 | 1.0 | 8.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 66.0 | 366.0 | 21.0 | 3.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 61.0 | 0.0 | 2.0 | 3.0 | 364.0 | 0.0 | 12.0 | 0.0 | 17.0 | 1.0 | 0.0 | 3.0 | 2.0 | 0.0 | 1.0 | 0.0 |
| 11 | 63.0 | 9.0 | 337.0 | 35.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 7.0 | 6.0 | 1.0 | 0.0 | 5.0 | 0.0 |
| 12 | 10.0 | 19.0 | 33.0 | 3.0 | 4.0 | 2.0 | 3.0 | 1.0 | 52.0 | 134.0 | 135.0 | 29.0 | 9.0 | 7.0 | 16.0 | 9.0 |
| 13 | 12.0 | 20.0 | 17.0 | 26.0 | 39.0 | 43.0 | 4.0 | 76.0 | 62.0 | 45.0 | 36.0 | 44.0 | 9.0 | 9.0 | 7.0 | 17.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.237 | -0.791 | 1.204 | -0.651 | -0.112 | -0.651 | -0.868 | -0.112 | 0.338 | -0.529 | 0.732 | -1.701 | -1.146 | -0.588 | 0.811 | -1.146 |
| 02 | -1.531 | -0.719 | 1.152 | -4.342 | -1.258 | -0.112 | -0.588 | -0.952 | -2.516 | 0.911 | 1.595 | -0.529 | -2.516 | -0.868 | 0.237 | -1.907 |
| 03 | -3.06 | -3.06 | -4.342 | -0.588 | -0.473 | -0.651 | -4.342 | 1.141 | -0.191 | -0.191 | -2.516 | 2.102 | -4.342 | -1.701 | -4.342 | -0.075 |
| 04 | -1.531 | -2.516 | 0.182 | -4.342 | -2.166 | -4.342 | 0.289 | -2.166 | -4.342 | -4.342 | -2.516 | -4.342 | -1.385 | -2.166 | 2.514 | -2.516 |
| 05 | -1.701 | -2.166 | -1.701 | -2.166 | -2.516 | -2.166 | -4.342 | -4.342 | 1.292 | 1.431 | 1.086 | 1.431 | -3.06 | -3.06 | -2.516 | -3.06 |
| 06 | -1.385 | -2.166 | -3.06 | 1.263 | -1.907 | -2.516 | -3.06 | 1.431 | -3.06 | -1.907 | -2.166 | 1.074 | -1.907 | -3.06 | -0.37 | 1.244 |
| 07 | -4.342 | -4.342 | -0.588 | -4.342 | -3.06 | -4.342 | -1.146 | -4.342 | -2.166 | -4.342 | -0.276 | -4.342 | -1.385 | -3.06 | 2.625 | -4.342 |
| 08 | -1.044 | -3.06 | -4.342 | -4.342 | -3.06 | -4.342 | -4.342 | -4.342 | 2.714 | -3.06 | -1.258 | -4.342 | -4.342 | -4.342 | -4.342 | -4.342 |
| 09 | 0.811 | 2.519 | -0.322 | -2.166 | -4.342 | -2.516 | -4.342 | -4.342 | -4.342 | -1.258 | -4.342 | -4.342 | -4.342 | -4.342 | -4.342 | -4.342 |
| 10 | 0.732 | -4.342 | -2.516 | -2.166 | 2.514 | -4.342 | -0.868 | -4.342 | -0.529 | -3.06 | -4.342 | -2.166 | -2.516 | -4.342 | -3.06 | -4.342 |
| 11 | 0.765 | -1.146 | 2.437 | 0.182 | -4.342 | -4.342 | -3.06 | -4.342 | -3.06 | -3.06 | -1.385 | -1.531 | -3.06 | -4.342 | -1.701 | -4.342 |
| 12 | -1.044 | -0.42 | 0.123 | -2.166 | -1.907 | -2.516 | -2.166 | -3.06 | 0.574 | 1.516 | 1.523 | -0.004 | -1.146 | -1.385 | -0.588 | -1.146 |
| 13 | -0.868 | -0.37 | -0.529 | -0.112 | 0.289 | 0.385 | -1.907 | 0.951 | 0.749 | 0.43 | 0.209 | 0.408 | -1.146 | -1.146 | -1.385 | -0.529 |