Model info
| Transcription factor | Tgif1 | ||||||||
| Model | TGIF1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 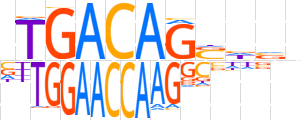 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 11 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nTGACAGSnvn | ||||||||
| Best auROC (human) | 0.812 | ||||||||
| Best auROC (mouse) | 0.981 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 5 | ||||||||
| Aligned words | 142 | ||||||||
| TF family | TALE-type homeo domain factors {3.1.4} | ||||||||
| TF subfamily | TGIF {3.1.4.6} | ||||||||
| MGI | MGI:1194497 | ||||||||
| EntrezGene | GeneID:21815 (SSTAR profile) | ||||||||
| UniProt ID | TGIF1_MOUSE | ||||||||
| UniProt AC | P70284 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Tgif1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.0 | 0.0 | 1.0 | 24.0 | 0.0 | 2.0 | 0.0 | 43.0 | 0.0 | 2.0 | 3.0 | 38.0 | 4.0 | 0.0 | 0.0 | 18.0 |
| 02 | 1.0 | 0.0 | 4.0 | 0.0 | 0.0 | 2.0 | 2.0 | 0.0 | 0.0 | 0.0 | 3.0 | 1.0 | 0.0 | 1.0 | 122.0 | 0.0 |
| 03 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 1.0 | 0.0 | 130.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 04 | 2.0 | 132.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 05 | 1.0 | 1.0 | 0.0 | 0.0 | 133.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 11.0 | 5.0 | 118.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 5.0 | 7.0 | 0.0 | 2.0 | 4.0 | 0.0 | 0.0 | 11.0 | 74.0 | 21.0 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 4.0 | 1.0 | 6.0 | 2.0 | 19.0 | 12.0 | 9.0 | 43.0 | 6.0 | 5.0 | 1.0 | 16.0 | 0.0 | 0.0 | 2.0 | 10.0 |
| 09 | 6.0 | 5.0 | 18.0 | 0.0 | 10.0 | 1.0 | 2.0 | 5.0 | 0.0 | 4.0 | 10.0 | 4.0 | 4.0 | 22.0 | 38.0 | 7.0 |
| 10 | 4.0 | 6.0 | 8.0 | 2.0 | 9.0 | 10.0 | 3.0 | 10.0 | 15.0 | 20.0 | 16.0 | 17.0 | 3.0 | 2.0 | 10.0 | 1.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.908 | -3.356 | -1.908 | 1.015 | -3.356 | -1.34 | -3.356 | 1.593 | -3.356 | -1.34 | -0.979 | 1.47 | -0.715 | -3.356 | -3.356 | 0.732 |
| 02 | -1.908 | -3.356 | -0.715 | -3.356 | -3.356 | -1.34 | -1.34 | -3.356 | -3.356 | -3.356 | -0.979 | -1.908 | -3.356 | -1.908 | 2.631 | -3.356 |
| 03 | -1.908 | -3.356 | -3.356 | -3.356 | -1.34 | -3.356 | -1.908 | -3.356 | 2.694 | -3.356 | -1.908 | -3.356 | -1.908 | -3.356 | -3.356 | -3.356 |
| 04 | -1.34 | 2.71 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -1.34 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 |
| 05 | -1.908 | -1.908 | -3.356 | -3.356 | 2.717 | -1.908 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 |
| 06 | 0.25 | -0.507 | 2.598 | -3.356 | -1.908 | -1.908 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 | -3.356 |
| 07 | -3.356 | -0.507 | -0.187 | -3.356 | -1.34 | -0.715 | -3.356 | -3.356 | 0.25 | 2.133 | 0.883 | 0.335 | -3.356 | -3.356 | -3.356 | -3.356 |
| 08 | -0.715 | -1.908 | -0.334 | -1.34 | 0.785 | 0.335 | 0.055 | 1.593 | -0.334 | -0.507 | -1.908 | 0.616 | -3.356 | -3.356 | -1.34 | 0.157 |
| 09 | -0.334 | -0.507 | 0.732 | -3.356 | 0.157 | -1.908 | -1.34 | -0.507 | -3.356 | -0.715 | 0.157 | -0.715 | -0.715 | 0.929 | 1.47 | -0.187 |
| 10 | -0.715 | -0.334 | -0.058 | -1.34 | 0.055 | 0.157 | -0.979 | 0.157 | 0.553 | 0.835 | 0.616 | 0.676 | -0.979 | -1.34 | 0.157 | -1.908 |