Model info
| Transcription factor | Znf281 | ||||||||
| Model | ZN281_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 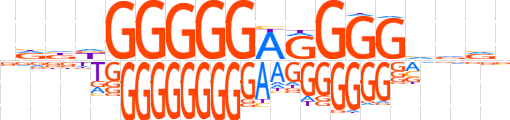 | ||||||||
| LOGO (reverse complement) | 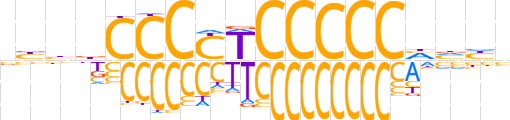 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vdbdGGGGGAGGGGvvvv | ||||||||
| Best auROC (human) | 0.942 | ||||||||
| Best auROC (mouse) | 0.999 | ||||||||
| Peak sets in benchmark (human) | 7 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 423 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | ZNF148-like factors {2.3.3.13} | ||||||||
| MGI | MGI:3029290 | ||||||||
| EntrezGene | GeneID:226442 (SSTAR profile) | ||||||||
| UniProt ID | ZN281_MOUSE | ||||||||
| UniProt AC | Q99LI5 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Znf281 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 21.0 | 4.0 | 39.0 | 7.0 | 15.0 | 12.0 | 32.0 | 19.0 | 50.0 | 20.0 | 127.0 | 20.0 | 5.0 | 4.0 | 37.0 | 6.0 |
| 02 | 12.0 | 7.0 | 61.0 | 11.0 | 5.0 | 3.0 | 20.0 | 12.0 | 36.0 | 48.0 | 108.0 | 43.0 | 2.0 | 4.0 | 35.0 | 11.0 |
| 03 | 10.0 | 5.0 | 13.0 | 27.0 | 2.0 | 4.0 | 18.0 | 38.0 | 51.0 | 29.0 | 7.0 | 137.0 | 6.0 | 2.0 | 36.0 | 33.0 |
| 04 | 0.0 | 0.0 | 69.0 | 0.0 | 0.0 | 0.0 | 40.0 | 0.0 | 1.0 | 0.0 | 73.0 | 0.0 | 0.0 | 0.0 | 234.0 | 1.0 |
| 05 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 415.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 06 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 411.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 1.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 412.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 416.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 339.0 | 37.0 | 1.0 | 40.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 38.0 | 0.0 | 264.0 | 37.0 | 7.0 | 0.0 | 23.0 | 7.0 | 0.0 | 0.0 | 1.0 | 0.0 | 3.0 | 0.0 | 36.0 | 2.0 |
| 11 | 0.0 | 0.0 | 48.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 322.0 | 0.0 | 0.0 | 0.0 | 46.0 | 0.0 |
| 12 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 17.0 | 3.0 | 385.0 | 11.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 0.0 | 2.0 | 15.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 16.0 | 5.0 | 359.0 | 7.0 | 2.0 | 0.0 | 9.0 | 0.0 |
| 14 | 4.0 | 4.0 | 10.0 | 0.0 | 2.0 | 0.0 | 2.0 | 3.0 | 157.0 | 104.0 | 82.0 | 41.0 | 3.0 | 1.0 | 5.0 | 0.0 |
| 15 | 45.0 | 24.0 | 88.0 | 9.0 | 34.0 | 21.0 | 41.0 | 13.0 | 35.0 | 39.0 | 12.0 | 13.0 | 4.0 | 7.0 | 27.0 | 6.0 |
| 16 | 23.0 | 16.0 | 68.0 | 11.0 | 22.0 | 14.0 | 41.0 | 14.0 | 38.0 | 34.0 | 85.0 | 11.0 | 7.0 | 4.0 | 26.0 | 4.0 |
| 17 | 14.0 | 14.0 | 52.0 | 10.0 | 8.0 | 19.0 | 23.0 | 18.0 | 37.0 | 71.0 | 89.0 | 23.0 | 4.0 | 6.0 | 19.0 | 11.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.215 | -1.801 | 0.396 | -1.279 | -0.544 | -0.761 | 0.2 | -0.313 | 0.642 | -0.263 | 1.57 | -0.263 | -1.595 | -1.801 | 0.344 | -1.424 |
| 02 | -0.761 | -1.279 | 0.84 | -0.846 | -1.595 | -2.06 | -0.263 | -0.761 | 0.317 | 0.602 | 1.408 | 0.493 | -2.411 | -1.801 | 0.289 | -0.846 |
| 03 | -0.938 | -1.595 | -0.684 | 0.032 | -2.411 | -1.801 | -0.366 | 0.37 | 0.662 | 0.103 | -1.279 | 1.646 | -1.424 | -2.411 | 0.317 | 0.231 |
| 04 | -4.252 | -4.252 | 0.962 | -4.252 | -4.252 | -4.252 | 0.421 | -4.252 | -2.957 | -4.252 | 1.018 | -4.252 | -4.252 | -4.252 | 2.18 | -2.957 |
| 05 | -4.252 | -4.252 | -2.957 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 | -2.957 | -4.252 | 2.752 | -4.252 | -4.252 | -4.252 | -2.957 | -4.252 |
| 06 | -4.252 | -4.252 | -2.957 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 | -1.424 | -4.252 | 2.742 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 |
| 07 | -2.957 | -4.252 | -1.595 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 | 2.745 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 |
| 08 | -4.252 | -4.252 | -2.957 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 | -2.957 | -4.252 | 2.754 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 |
| 09 | -4.252 | -4.252 | -4.252 | -2.957 | -4.252 | -4.252 | -4.252 | -4.252 | 2.55 | 0.344 | -2.957 | 0.421 | -4.252 | -4.252 | -4.252 | -4.252 |
| 10 | 0.37 | -4.252 | 2.3 | 0.344 | -1.279 | -4.252 | -0.125 | -1.279 | -4.252 | -4.252 | -2.957 | -4.252 | -2.06 | -4.252 | 0.317 | -2.411 |
| 11 | -4.252 | -4.252 | 0.602 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 | -2.411 | -4.252 | 2.498 | -4.252 | -4.252 | -4.252 | 0.56 | -4.252 |
| 12 | -4.252 | -4.252 | -2.411 | -4.252 | -4.252 | -4.252 | -4.252 | -4.252 | -0.422 | -2.06 | 2.677 | -0.846 | -4.252 | -4.252 | -4.252 | -4.252 |
| 13 | -4.252 | -2.411 | -0.544 | -4.252 | -4.252 | -4.252 | -2.957 | -2.411 | -0.481 | -1.595 | 2.607 | -1.279 | -2.411 | -4.252 | -1.039 | -4.252 |
| 14 | -1.801 | -1.801 | -0.938 | -4.252 | -2.411 | -4.252 | -2.411 | -2.06 | 1.781 | 1.371 | 1.134 | 0.446 | -2.06 | -2.957 | -1.595 | -4.252 |
| 15 | 0.538 | -0.084 | 1.204 | -1.039 | 0.26 | -0.215 | 0.446 | -0.684 | 0.289 | 0.396 | -0.761 | -0.684 | -1.801 | -1.279 | 0.032 | -1.424 |
| 16 | -0.125 | -0.481 | 0.948 | -0.846 | -0.169 | -0.612 | 0.446 | -0.612 | 0.37 | 0.26 | 1.17 | -0.846 | -1.279 | -1.801 | -0.005 | -1.801 |
| 17 | -0.612 | -0.612 | 0.681 | -0.938 | -1.152 | -0.313 | -0.125 | -0.366 | 0.344 | 0.991 | 1.216 | -0.125 | -1.801 | -1.424 | -0.313 | -0.846 |