Model info
| Transcription factor | ZNF394 (GeneCards) | ||||||||
| Model | ZN394_HUMAN.H11MO.1.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 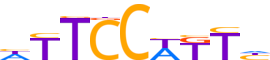 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 9 | ||||||||
| Quality | D | ||||||||
| Motif rank | 1 | ||||||||
| Consensus | dAMWGGARd | ||||||||
| Best auROC (human) | 0.661 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 382 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:18832 | ||||||||
| EntrezGene | GeneID:84124 (SSTAR profile) | ||||||||
| UniProt ID | ZN394_HUMAN | ||||||||
| UniProt AC | Q53GI3 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF394 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF394 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 79.0 | 34.0 | 202.0 | 64.0 |
| 02 | 299.0 | 10.0 | 69.0 | 1.0 |
| 03 | 261.0 | 90.0 | 24.0 | 4.0 |
| 04 | 155.0 | 2.0 | 18.0 | 204.0 |
| 05 | 6.0 | 0.0 | 373.0 | 0.0 |
| 06 | 11.0 | 13.0 | 347.0 | 8.0 |
| 07 | 351.0 | 12.0 | 11.0 | 5.0 |
| 08 | 289.0 | 13.0 | 68.0 | 9.0 |
| 09 | 107.0 | 13.0 | 70.0 | 189.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.179 | -0.998 | 0.749 | -0.385 |
| 02 | 1.139 | -2.126 | -0.311 | -3.657 |
| 03 | 1.003 | -0.051 | -1.329 | -2.865 |
| 04 | 0.486 | -3.318 | -1.597 | 0.759 |
| 05 | -2.554 | -4.172 | 1.359 | -4.172 |
| 06 | -2.042 | -1.894 | 1.287 | -2.317 |
| 07 | 1.298 | -1.965 | -2.042 | -2.697 |
| 08 | 1.105 | -1.894 | -0.326 | -2.217 |
| 09 | 0.12 | -1.894 | -0.297 | 0.683 |