Model info
| Transcription factor | ARNT (GeneCards) | ||||||||
| Model | ARNT_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 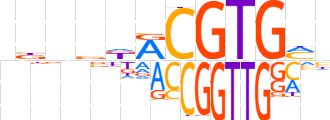 | ||||||||
| LOGO (reverse complement) | 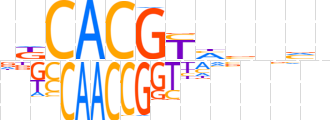 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbnvdRCGTGMb | ||||||||
| Best auROC (human) | 0.952 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 8 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 512 | ||||||||
| TF family | PAS domain factors {1.2.5} | ||||||||
| TF subfamily | Arnt-like factors {1.2.5.2} | ||||||||
| HGNC | HGNC:700 | ||||||||
| EntrezGene | GeneID:405 (SSTAR profile) | ||||||||
| UniProt ID | ARNT_HUMAN | ||||||||
| UniProt AC | P27540 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ARNT expression | ||||||||
| ReMap ChIP-seq dataset list | ARNT datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 13.0 | 13.0 | 38.0 | 16.0 | 16.0 | 21.0 | 38.0 | 37.0 | 27.0 | 29.0 | 84.0 | 40.0 | 6.0 | 12.0 | 92.0 | 18.0 |
| 02 | 13.0 | 11.0 | 23.0 | 15.0 | 23.0 | 13.0 | 18.0 | 21.0 | 46.0 | 82.0 | 68.0 | 56.0 | 13.0 | 6.0 | 82.0 | 10.0 |
| 03 | 21.0 | 19.0 | 39.0 | 16.0 | 14.0 | 44.0 | 33.0 | 21.0 | 40.0 | 68.0 | 71.0 | 12.0 | 10.0 | 11.0 | 70.0 | 11.0 |
| 04 | 23.0 | 9.0 | 31.0 | 22.0 | 16.0 | 10.0 | 9.0 | 107.0 | 23.0 | 14.0 | 65.0 | 111.0 | 15.0 | 5.0 | 9.0 | 31.0 |
| 05 | 56.0 | 0.0 | 18.0 | 3.0 | 26.0 | 4.0 | 7.0 | 1.0 | 99.0 | 6.0 | 8.0 | 1.0 | 180.0 | 9.0 | 77.0 | 5.0 |
| 06 | 6.0 | 348.0 | 1.0 | 6.0 | 0.0 | 17.0 | 0.0 | 2.0 | 0.0 | 108.0 | 0.0 | 2.0 | 0.0 | 10.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 6.0 | 0.0 | 2.0 | 1.0 | 480.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 10.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 497.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 498.0 | 1.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 172.0 | 254.0 | 19.0 | 53.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 11 | 8.0 | 116.0 | 32.0 | 17.0 | 63.0 | 84.0 | 38.0 | 69.0 | 4.0 | 2.0 | 11.0 | 2.0 | 3.0 | 16.0 | 23.0 | 12.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.86 | -0.86 | 0.193 | -0.658 | -0.658 | -0.392 | 0.193 | 0.167 | -0.144 | -0.074 | 0.981 | 0.244 | -1.6 | -0.938 | 1.072 | -0.543 |
| 02 | -0.86 | -1.022 | -0.302 | -0.721 | -0.302 | -0.86 | -0.543 | -0.392 | 0.383 | 0.957 | 0.771 | 0.578 | -0.86 | -1.6 | 0.957 | -1.114 |
| 03 | -0.392 | -0.49 | 0.219 | -0.658 | -0.788 | 0.339 | 0.054 | -0.392 | 0.244 | 0.771 | 0.814 | -0.938 | -1.114 | -1.022 | 0.8 | -1.022 |
| 04 | -0.302 | -1.215 | -0.008 | -0.346 | -0.658 | -1.114 | -1.215 | 1.222 | -0.302 | -0.788 | 0.726 | 1.259 | -0.721 | -1.77 | -1.215 | -0.008 |
| 05 | 0.578 | -4.4 | -0.543 | -2.234 | -0.181 | -1.975 | -1.454 | -3.126 | 1.145 | -1.6 | -1.328 | -3.126 | 1.741 | -1.215 | 0.894 | -1.77 |
| 06 | -1.6 | 2.399 | -3.126 | -1.6 | -4.4 | -0.599 | -4.4 | -2.584 | -4.4 | 1.231 | -4.4 | -2.584 | -4.4 | -1.114 | -4.4 | -4.4 |
| 07 | -4.4 | -4.4 | -1.6 | -4.4 | -2.584 | -3.126 | 2.72 | -4.4 | -4.4 | -4.4 | -3.126 | -4.4 | -4.4 | -4.4 | -1.114 | -4.4 |
| 08 | -4.4 | -4.4 | -4.4 | -2.584 | -4.4 | -4.4 | -4.4 | -3.126 | -4.4 | -4.4 | -4.4 | 2.755 | -4.4 | -4.4 | -4.4 | -4.4 |
| 09 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.126 | 2.757 | -3.126 |
| 10 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.126 | 1.695 | 2.084 | -0.49 | 0.523 | -3.126 | -4.4 | -4.4 | -4.4 |
| 11 | -1.328 | 1.303 | 0.023 | -0.599 | 0.695 | 0.981 | 0.193 | 0.785 | -1.975 | -2.584 | -1.022 | -2.584 | -2.234 | -0.658 | -0.302 | -0.938 |