Model info
| Transcription factor | CTCFL (GeneCards) | ||||||||
| Model | CTCFL_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 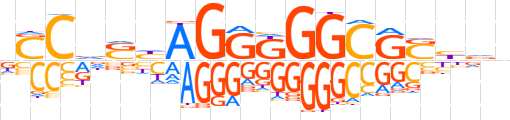 | ||||||||
| LOGO (reverse complement) | 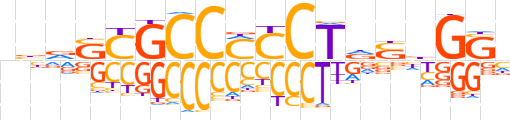 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vSCdShAGGKGGCGShnn | ||||||||
| Best auROC (human) | 0.98 | ||||||||
| Best auROC (mouse) | 0.995 | ||||||||
| Peak sets in benchmark (human) | 25 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 490 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | CTCF-like factors {2.3.3.50} | ||||||||
| HGNC | HGNC:16234 | ||||||||
| EntrezGene | GeneID:140690 (SSTAR profile) | ||||||||
| UniProt ID | CTCFL_HUMAN | ||||||||
| UniProt AC | Q8NI51 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | CTCFL expression | ||||||||
| ReMap ChIP-seq dataset list | CTCFL datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 9.0 | 68.0 | 11.0 | 6.0 | 4.0 | 66.0 | 15.0 | 6.0 | 15.0 | 167.0 | 17.0 | 10.0 | 1.0 | 79.0 | 10.0 | 1.0 |
| 02 | 2.0 | 26.0 | 0.0 | 1.0 | 9.0 | 366.0 | 2.0 | 3.0 | 2.0 | 41.0 | 7.0 | 3.0 | 1.0 | 21.0 | 1.0 | 0.0 |
| 03 | 5.0 | 0.0 | 8.0 | 1.0 | 176.0 | 39.0 | 140.0 | 99.0 | 3.0 | 3.0 | 4.0 | 0.0 | 0.0 | 3.0 | 3.0 | 1.0 |
| 04 | 13.0 | 90.0 | 80.0 | 1.0 | 8.0 | 19.0 | 16.0 | 2.0 | 17.0 | 97.0 | 39.0 | 2.0 | 6.0 | 31.0 | 64.0 | 0.0 |
| 05 | 5.0 | 24.0 | 8.0 | 7.0 | 34.0 | 107.0 | 13.0 | 83.0 | 27.0 | 117.0 | 8.0 | 47.0 | 1.0 | 1.0 | 3.0 | 0.0 |
| 06 | 49.0 | 6.0 | 7.0 | 5.0 | 235.0 | 3.0 | 0.0 | 11.0 | 20.0 | 7.0 | 1.0 | 4.0 | 122.0 | 2.0 | 8.0 | 5.0 |
| 07 | 3.0 | 0.0 | 421.0 | 2.0 | 0.0 | 0.0 | 18.0 | 0.0 | 0.0 | 2.0 | 14.0 | 0.0 | 0.0 | 0.0 | 25.0 | 0.0 |
| 08 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 103.0 | 3.0 | 369.0 | 3.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| 09 | 1.0 | 4.0 | 96.0 | 3.0 | 1.0 | 2.0 | 1.0 | 0.0 | 33.0 | 24.0 | 272.0 | 44.0 | 0.0 | 1.0 | 3.0 | 0.0 |
| 10 | 1.0 | 1.0 | 33.0 | 0.0 | 2.0 | 0.0 | 29.0 | 0.0 | 2.0 | 3.0 | 365.0 | 2.0 | 0.0 | 0.0 | 47.0 | 0.0 |
| 11 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 1.0 | 1.0 | 2.0 | 3.0 | 6.0 | 447.0 | 18.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 12 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 42.0 | 398.0 | 4.0 | 11.0 | 1.0 | 18.0 | 0.0 | 1.0 |
| 13 | 2.0 | 1.0 | 40.0 | 0.0 | 109.0 | 2.0 | 308.0 | 7.0 | 0.0 | 0.0 | 4.0 | 0.0 | 1.0 | 1.0 | 10.0 | 0.0 |
| 14 | 1.0 | 22.0 | 86.0 | 3.0 | 3.0 | 1.0 | 0.0 | 0.0 | 17.0 | 301.0 | 27.0 | 17.0 | 1.0 | 6.0 | 0.0 | 0.0 |
| 15 | 0.0 | 10.0 | 3.0 | 9.0 | 24.0 | 126.0 | 26.0 | 154.0 | 21.0 | 48.0 | 7.0 | 37.0 | 4.0 | 7.0 | 3.0 | 6.0 |
| 16 | 4.0 | 17.0 | 19.0 | 9.0 | 91.0 | 56.0 | 25.0 | 19.0 | 2.0 | 21.0 | 14.0 | 2.0 | 37.0 | 40.0 | 120.0 | 9.0 |
| 17 | 21.0 | 41.0 | 55.0 | 17.0 | 16.0 | 16.0 | 40.0 | 62.0 | 20.0 | 74.0 | 31.0 | 53.0 | 3.0 | 10.0 | 16.0 | 10.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.185 | 0.801 | -0.992 | -1.57 | -1.946 | 0.771 | -0.691 | -1.57 | -0.691 | 1.696 | -0.569 | -1.084 | -3.097 | 0.95 | -1.084 | -3.097 |
| 02 | -2.554 | -0.151 | -4.375 | -3.097 | -1.185 | 2.479 | -2.554 | -2.204 | -2.554 | 0.299 | -1.425 | -2.204 | -3.097 | -0.361 | -3.097 | -4.375 |
| 03 | -1.74 | -4.375 | -1.298 | -3.097 | 1.748 | 0.249 | 1.52 | 1.175 | -2.204 | -2.204 | -1.946 | -4.375 | -4.375 | -2.204 | -2.204 | -3.097 |
| 04 | -0.83 | 1.08 | 0.963 | -3.097 | -1.298 | -0.46 | -0.628 | -2.554 | -0.569 | 1.154 | 0.249 | -2.554 | -1.57 | 0.022 | 0.741 | -4.375 |
| 05 | -1.74 | -0.23 | -1.298 | -1.425 | 0.113 | 1.252 | -0.83 | 0.999 | -0.114 | 1.341 | -1.298 | 0.434 | -3.097 | -3.097 | -2.204 | -4.375 |
| 06 | 0.475 | -1.57 | -1.425 | -1.74 | 2.037 | -2.204 | -4.375 | -0.992 | -0.409 | -1.425 | -3.097 | -1.946 | 1.383 | -2.554 | -1.298 | -1.74 |
| 07 | -2.204 | -4.375 | 2.619 | -2.554 | -4.375 | -4.375 | -0.513 | -4.375 | -4.375 | -2.554 | -0.758 | -4.375 | -4.375 | -4.375 | -0.19 | -4.375 |
| 08 | -4.375 | -4.375 | -2.204 | -4.375 | -4.375 | -3.097 | -4.375 | -3.097 | 1.214 | -2.204 | 2.488 | -2.204 | -3.097 | -4.375 | -3.097 | -4.375 |
| 09 | -3.097 | -1.946 | 1.144 | -2.204 | -3.097 | -2.554 | -3.097 | -4.375 | 0.084 | -0.23 | 2.183 | 0.369 | -4.375 | -3.097 | -2.204 | -4.375 |
| 10 | -3.097 | -3.097 | 0.084 | -4.375 | -2.554 | -4.375 | -0.044 | -4.375 | -2.554 | -2.204 | 2.477 | -2.554 | -4.375 | -4.375 | 0.434 | -4.375 |
| 11 | -4.375 | -4.375 | -1.74 | -4.375 | -4.375 | -3.097 | -3.097 | -2.554 | -2.204 | -1.57 | 2.679 | -0.513 | -4.375 | -4.375 | -2.554 | -4.375 |
| 12 | -4.375 | -2.204 | -4.375 | -4.375 | -4.375 | -1.425 | -4.375 | -4.375 | 0.323 | 2.563 | -1.946 | -0.992 | -3.097 | -0.513 | -4.375 | -3.097 |
| 13 | -2.554 | -3.097 | 0.274 | -4.375 | 1.271 | -2.554 | 2.307 | -1.425 | -4.375 | -4.375 | -1.946 | -4.375 | -3.097 | -3.097 | -1.084 | -4.375 |
| 14 | -3.097 | -0.316 | 1.035 | -2.204 | -2.204 | -3.097 | -4.375 | -4.375 | -0.569 | 2.284 | -0.114 | -0.569 | -3.097 | -1.57 | -4.375 | -4.375 |
| 15 | -4.375 | -1.084 | -2.204 | -1.185 | -0.23 | 1.415 | -0.151 | 1.615 | -0.361 | 0.455 | -1.425 | 0.197 | -1.946 | -1.425 | -2.204 | -1.57 |
| 16 | -1.946 | -0.569 | -0.46 | -1.185 | 1.091 | 0.608 | -0.19 | -0.46 | -2.554 | -0.361 | -0.758 | -2.554 | 0.197 | 0.274 | 1.366 | -1.185 |
| 17 | -0.361 | 0.299 | 0.59 | -0.569 | -0.628 | -0.628 | 0.274 | 0.709 | -0.409 | 0.885 | 0.022 | 0.553 | -2.204 | -1.084 | -0.628 | -1.084 |