Model info
| Transcription factor | GATA4 (GeneCards) | ||||||||
| Model | GATA4_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 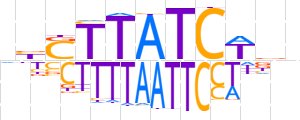 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 11 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvWGATAASvn | ||||||||
| Best auROC (human) | 0.96 | ||||||||
| Best auROC (mouse) | 0.945 | ||||||||
| Peak sets in benchmark (human) | 15 | ||||||||
| Peak sets in benchmark (mouse) | 17 | ||||||||
| Aligned words | 463 | ||||||||
| TF family | GATA-type zinc fingers {2.2.1} | ||||||||
| TF subfamily | Two zinc-finger GATA factors {2.2.1.1} | ||||||||
| HGNC | HGNC:4173 | ||||||||
| EntrezGene | GeneID:2626 (SSTAR profile) | ||||||||
| UniProt ID | GATA4_HUMAN | ||||||||
| UniProt AC | P43694 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | GATA4 expression | ||||||||
| ReMap ChIP-seq dataset list | GATA4 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 31.0 | 61.0 | 45.0 | 15.0 | 24.0 | 30.0 | 4.0 | 18.0 | 19.0 | 40.0 | 23.0 | 16.0 | 34.0 | 48.0 | 23.0 | 14.0 |
| 02 | 79.0 | 5.0 | 2.0 | 22.0 | 85.0 | 4.0 | 1.0 | 89.0 | 68.0 | 5.0 | 0.0 | 22.0 | 42.0 | 2.0 | 1.0 | 18.0 |
| 03 | 3.0 | 0.0 | 268.0 | 3.0 | 4.0 | 0.0 | 12.0 | 0.0 | 0.0 | 1.0 | 3.0 | 0.0 | 2.0 | 0.0 | 148.0 | 1.0 |
| 04 | 8.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 417.0 | 4.0 | 5.0 | 5.0 | 4.0 | 0.0 | 0.0 | 0.0 |
| 05 | 10.0 | 6.0 | 8.0 | 406.0 | 1.0 | 1.0 | 0.0 | 2.0 | 0.0 | 0.0 | 2.0 | 3.0 | 1.0 | 0.0 | 0.0 | 5.0 |
| 06 | 9.0 | 1.0 | 0.0 | 2.0 | 5.0 | 0.0 | 0.0 | 2.0 | 10.0 | 0.0 | 0.0 | 0.0 | 386.0 | 5.0 | 2.0 | 23.0 |
| 07 | 367.0 | 7.0 | 22.0 | 14.0 | 3.0 | 0.0 | 1.0 | 2.0 | 1.0 | 0.0 | 1.0 | 0.0 | 20.0 | 0.0 | 7.0 | 0.0 |
| 08 | 34.0 | 59.0 | 293.0 | 5.0 | 0.0 | 2.0 | 4.0 | 1.0 | 0.0 | 14.0 | 15.0 | 2.0 | 1.0 | 4.0 | 10.0 | 1.0 |
| 09 | 9.0 | 3.0 | 20.0 | 3.0 | 56.0 | 8.0 | 5.0 | 10.0 | 164.0 | 53.0 | 83.0 | 22.0 | 2.0 | 1.0 | 5.0 | 1.0 |
| 10 | 66.0 | 28.0 | 58.0 | 79.0 | 27.0 | 18.0 | 1.0 | 19.0 | 42.0 | 18.0 | 28.0 | 25.0 | 11.0 | 6.0 | 7.0 | 12.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.107 | 0.778 | 0.476 | -0.606 | -0.145 | 0.075 | -1.862 | -0.428 | -0.375 | 0.359 | -0.187 | -0.543 | 0.198 | 0.54 | -0.187 | -0.673 |
| 02 | 1.035 | -1.656 | -2.472 | -0.231 | 1.108 | -1.862 | -3.016 | 1.154 | 0.886 | -1.656 | -4.304 | -0.231 | 0.408 | -2.472 | -3.016 | -0.428 |
| 03 | -2.121 | -4.304 | 2.253 | -2.121 | -1.862 | -4.304 | -0.823 | -4.304 | -4.304 | -3.016 | -2.121 | -4.304 | -2.472 | -4.304 | 1.661 | -3.016 |
| 04 | -1.213 | -4.304 | -4.304 | -3.016 | -3.016 | -4.304 | -4.304 | -4.304 | 2.695 | -1.862 | -1.656 | -1.656 | -1.862 | -4.304 | -4.304 | -4.304 |
| 05 | -0.999 | -1.486 | -1.213 | 2.668 | -3.016 | -3.016 | -4.304 | -2.472 | -4.304 | -4.304 | -2.472 | -2.121 | -3.016 | -4.304 | -4.304 | -1.656 |
| 06 | -1.1 | -3.016 | -4.304 | -2.472 | -1.656 | -4.304 | -4.304 | -2.472 | -0.999 | -4.304 | -4.304 | -4.304 | 2.618 | -1.656 | -2.472 | -0.187 |
| 07 | 2.567 | -1.34 | -0.231 | -0.673 | -2.121 | -4.304 | -3.016 | -2.472 | -3.016 | -4.304 | -3.016 | -4.304 | -0.324 | -4.304 | -1.34 | -4.304 |
| 08 | 0.198 | 0.745 | 2.342 | -1.656 | -4.304 | -2.472 | -1.862 | -3.016 | -4.304 | -0.673 | -0.606 | -2.472 | -3.016 | -1.862 | -0.999 | -3.016 |
| 09 | -1.1 | -2.121 | -0.324 | -2.121 | 0.693 | -1.213 | -1.656 | -0.999 | 1.763 | 0.638 | 1.084 | -0.231 | -2.472 | -3.016 | -1.656 | -3.016 |
| 10 | 0.856 | 0.007 | 0.728 | 1.035 | -0.029 | -0.428 | -3.016 | -0.375 | 0.408 | -0.428 | 0.007 | -0.105 | -0.907 | -1.486 | -1.34 | -0.823 |