Model info
| Transcription factor | GATA6 (GeneCards) | ||||||||
| Model | GATA6_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 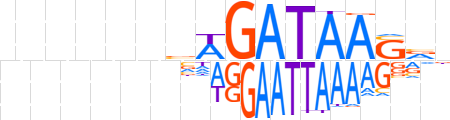 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnnnnnvWGATAASvn | ||||||||
| Best auROC (human) | 0.977 | ||||||||
| Best auROC (mouse) | 0.946 | ||||||||
| Peak sets in benchmark (human) | 29 | ||||||||
| Peak sets in benchmark (mouse) | 7 | ||||||||
| Aligned words | 394 | ||||||||
| TF family | GATA-type zinc fingers {2.2.1} | ||||||||
| TF subfamily | Two zinc-finger GATA factors {2.2.1.1} | ||||||||
| HGNC | HGNC:4174 | ||||||||
| EntrezGene | GeneID:2627 (SSTAR profile) | ||||||||
| UniProt ID | GATA6_HUMAN | ||||||||
| UniProt AC | Q92908 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | GATA6 expression | ||||||||
| ReMap ChIP-seq dataset list | GATA6 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 22.0 | 16.0 | 21.0 | 26.0 | 41.0 | 39.0 | 4.0 | 10.0 | 27.0 | 57.0 | 30.0 | 11.0 | 14.0 | 27.0 | 34.0 | 11.0 |
| 02 | 38.0 | 22.0 | 24.0 | 20.0 | 45.0 | 40.0 | 6.0 | 48.0 | 19.0 | 25.0 | 29.0 | 16.0 | 12.0 | 23.0 | 15.0 | 8.0 |
| 03 | 29.0 | 22.0 | 46.0 | 17.0 | 43.0 | 34.0 | 5.0 | 28.0 | 10.0 | 24.0 | 19.0 | 21.0 | 7.0 | 22.0 | 38.0 | 25.0 |
| 04 | 23.0 | 20.0 | 30.0 | 16.0 | 36.0 | 29.0 | 5.0 | 32.0 | 30.0 | 31.0 | 27.0 | 20.0 | 12.0 | 20.0 | 25.0 | 34.0 |
| 05 | 26.0 | 12.0 | 32.0 | 31.0 | 43.0 | 23.0 | 8.0 | 26.0 | 24.0 | 15.0 | 33.0 | 15.0 | 20.0 | 22.0 | 32.0 | 28.0 |
| 06 | 15.0 | 44.0 | 43.0 | 11.0 | 22.0 | 37.0 | 3.0 | 10.0 | 16.0 | 46.0 | 35.0 | 8.0 | 29.0 | 32.0 | 24.0 | 15.0 |
| 07 | 69.0 | 0.0 | 3.0 | 10.0 | 56.0 | 4.0 | 0.0 | 99.0 | 76.0 | 3.0 | 0.0 | 26.0 | 25.0 | 0.0 | 0.0 | 19.0 |
| 08 | 1.0 | 0.0 | 225.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 1.0 | 153.0 | 0.0 |
| 09 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 382.0 | 1.0 | 2.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 4.0 | 2.0 | 2.0 | 375.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 3.0 |
| 11 | 3.0 | 0.0 | 1.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 358.0 | 1.0 | 0.0 | 22.0 |
| 12 | 343.0 | 3.0 | 11.0 | 7.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 18.0 | 0.0 | 3.0 | 1.0 |
| 13 | 32.0 | 52.0 | 277.0 | 4.0 | 1.0 | 0.0 | 1.0 | 1.0 | 2.0 | 5.0 | 6.0 | 1.0 | 0.0 | 2.0 | 4.0 | 2.0 |
| 14 | 15.0 | 7.0 | 10.0 | 3.0 | 37.0 | 8.0 | 10.0 | 4.0 | 123.0 | 54.0 | 105.0 | 6.0 | 2.0 | 0.0 | 4.0 | 2.0 |
| 15 | 30.0 | 20.0 | 66.0 | 61.0 | 21.0 | 18.0 | 0.0 | 30.0 | 49.0 | 27.0 | 35.0 | 18.0 | 7.0 | 1.0 | 2.0 | 5.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.101 | -0.413 | -0.147 | 0.064 | 0.514 | 0.464 | -1.733 | -0.87 | 0.101 | 0.841 | 0.205 | -0.778 | -0.543 | 0.101 | 0.329 | -0.778 |
| 02 | 0.439 | -0.101 | -0.015 | -0.195 | 0.606 | 0.489 | -1.357 | 0.67 | -0.245 | 0.025 | 0.171 | -0.413 | -0.693 | -0.057 | -0.476 | -1.084 |
| 03 | 0.171 | -0.101 | 0.628 | -0.354 | 0.561 | 0.329 | -1.527 | 0.137 | -0.87 | -0.015 | -0.245 | -0.147 | -1.211 | -0.101 | 0.439 | 0.025 |
| 04 | -0.057 | -0.195 | 0.205 | -0.413 | 0.385 | 0.171 | -1.527 | 0.269 | 0.205 | 0.237 | 0.101 | -0.195 | -0.693 | -0.195 | 0.025 | 0.329 |
| 05 | 0.064 | -0.693 | 0.269 | 0.237 | 0.561 | -0.057 | -1.084 | 0.064 | -0.015 | -0.476 | 0.299 | -0.476 | -0.195 | -0.101 | 0.269 | 0.137 |
| 06 | -0.476 | 0.584 | 0.561 | -0.778 | -0.101 | 0.412 | -1.993 | -0.87 | -0.413 | 0.628 | 0.357 | -1.084 | 0.171 | 0.269 | -0.015 | -0.476 |
| 07 | 1.031 | -4.195 | -1.993 | -0.87 | 0.823 | -1.733 | -4.195 | 1.39 | 1.127 | -1.993 | -4.195 | 0.064 | 0.025 | -4.195 | -4.195 | -0.245 |
| 08 | -2.892 | -4.195 | 2.209 | -4.195 | -4.195 | -4.195 | -1.211 | -4.195 | -4.195 | -4.195 | -1.993 | -4.195 | -4.195 | -2.892 | 1.824 | -4.195 |
| 09 | -2.892 | -4.195 | -4.195 | -4.195 | -4.195 | -2.892 | -4.195 | -4.195 | 2.738 | -2.892 | -2.345 | -1.993 | -4.195 | -4.195 | -4.195 | -4.195 |
| 10 | -1.733 | -2.345 | -2.345 | 2.719 | -4.195 | -2.892 | -4.195 | -2.892 | -4.195 | -4.195 | -4.195 | -2.345 | -4.195 | -4.195 | -4.195 | -1.993 |
| 11 | -1.993 | -4.195 | -2.892 | -4.195 | -1.993 | -4.195 | -4.195 | -4.195 | -4.195 | -2.345 | -4.195 | -4.195 | 2.673 | -2.892 | -4.195 | -0.101 |
| 12 | 2.63 | -1.993 | -0.778 | -1.211 | -1.993 | -4.195 | -4.195 | -4.195 | -2.892 | -4.195 | -4.195 | -4.195 | -0.298 | -4.195 | -1.993 | -2.892 |
| 13 | 0.269 | 0.75 | 2.417 | -1.733 | -2.892 | -4.195 | -2.892 | -2.892 | -2.345 | -1.527 | -1.357 | -2.892 | -4.195 | -2.345 | -1.733 | -2.345 |
| 14 | -0.476 | -1.211 | -0.87 | -1.993 | 0.412 | -1.084 | -0.87 | -1.733 | 1.606 | 0.787 | 1.449 | -1.357 | -2.345 | -4.195 | -1.733 | -2.345 |
| 15 | 0.205 | -0.195 | 0.987 | 0.908 | -0.147 | -0.298 | -4.195 | 0.205 | 0.691 | 0.101 | 0.357 | -0.298 | -1.211 | -2.892 | -2.345 | -1.527 |