Model info
| Transcription factor | KLF5 (GeneCards) | ||||||||
| Model | KLF5_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 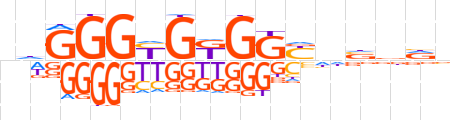 | ||||||||
| LOGO (reverse complement) | 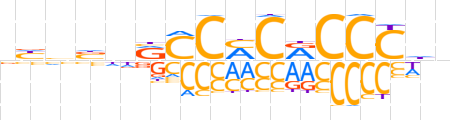 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndGGGYGKGKYnvddn | ||||||||
| Best auROC (human) | 0.973 | ||||||||
| Best auROC (mouse) | 0.978 | ||||||||
| Peak sets in benchmark (human) | 32 | ||||||||
| Peak sets in benchmark (mouse) | 23 | ||||||||
| Aligned words | 514 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | Krüppel-like factors {2.3.1.2} | ||||||||
| HGNC | HGNC:6349 | ||||||||
| EntrezGene | GeneID:688 (SSTAR profile) | ||||||||
| UniProt ID | KLF5_HUMAN | ||||||||
| UniProt AC | Q13887 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | KLF5 expression | ||||||||
| ReMap ChIP-seq dataset list | KLF5 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 38.0 | 9.0 | 36.0 | 24.0 | 67.0 | 15.0 | 22.0 | 63.0 | 66.0 | 17.0 | 19.0 | 40.0 | 21.0 | 7.0 | 19.0 | 25.0 |
| 02 | 16.0 | 2.0 | 172.0 | 2.0 | 19.0 | 0.0 | 22.0 | 7.0 | 13.0 | 0.0 | 79.0 | 4.0 | 17.0 | 2.0 | 131.0 | 2.0 |
| 03 | 3.0 | 0.0 | 61.0 | 1.0 | 0.0 | 0.0 | 3.0 | 1.0 | 4.0 | 7.0 | 382.0 | 11.0 | 2.0 | 3.0 | 7.0 | 3.0 |
| 04 | 1.0 | 0.0 | 7.0 | 1.0 | 1.0 | 1.0 | 6.0 | 2.0 | 7.0 | 5.0 | 435.0 | 6.0 | 0.0 | 3.0 | 12.0 | 1.0 |
| 05 | 1.0 | 4.0 | 1.0 | 3.0 | 0.0 | 0.0 | 0.0 | 9.0 | 47.0 | 134.0 | 3.0 | 276.0 | 2.0 | 3.0 | 2.0 | 3.0 |
| 06 | 5.0 | 0.0 | 44.0 | 1.0 | 1.0 | 4.0 | 126.0 | 10.0 | 0.0 | 1.0 | 4.0 | 1.0 | 5.0 | 3.0 | 278.0 | 5.0 |
| 07 | 1.0 | 0.0 | 4.0 | 6.0 | 1.0 | 0.0 | 1.0 | 6.0 | 25.0 | 7.0 | 176.0 | 244.0 | 1.0 | 0.0 | 11.0 | 5.0 |
| 08 | 1.0 | 1.0 | 24.0 | 2.0 | 0.0 | 0.0 | 6.0 | 1.0 | 4.0 | 3.0 | 180.0 | 5.0 | 5.0 | 5.0 | 245.0 | 6.0 |
| 09 | 1.0 | 0.0 | 7.0 | 2.0 | 1.0 | 3.0 | 4.0 | 1.0 | 10.0 | 11.0 | 353.0 | 81.0 | 0.0 | 0.0 | 11.0 | 3.0 |
| 10 | 0.0 | 9.0 | 1.0 | 2.0 | 3.0 | 11.0 | 0.0 | 0.0 | 38.0 | 252.0 | 34.0 | 51.0 | 6.0 | 54.0 | 20.0 | 7.0 |
| 11 | 16.0 | 12.0 | 13.0 | 6.0 | 116.0 | 65.0 | 33.0 | 112.0 | 18.0 | 4.0 | 7.0 | 26.0 | 13.0 | 15.0 | 19.0 | 13.0 |
| 12 | 13.0 | 21.0 | 105.0 | 24.0 | 32.0 | 21.0 | 27.0 | 16.0 | 10.0 | 19.0 | 39.0 | 4.0 | 10.0 | 26.0 | 101.0 | 20.0 |
| 13 | 11.0 | 8.0 | 37.0 | 9.0 | 32.0 | 12.0 | 11.0 | 32.0 | 51.0 | 38.0 | 125.0 | 58.0 | 6.0 | 14.0 | 26.0 | 18.0 |
| 14 | 20.0 | 7.0 | 62.0 | 11.0 | 18.0 | 10.0 | 26.0 | 18.0 | 50.0 | 19.0 | 115.0 | 15.0 | 13.0 | 17.0 | 74.0 | 13.0 |
| 15 | 30.0 | 27.0 | 34.0 | 10.0 | 24.0 | 17.0 | 3.0 | 9.0 | 54.0 | 78.0 | 95.0 | 50.0 | 9.0 | 17.0 | 27.0 | 4.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.217 | -1.191 | 0.164 | -0.236 | 0.78 | -0.697 | -0.322 | 0.719 | 0.765 | -0.575 | -0.466 | 0.268 | -0.368 | -1.431 | -0.466 | -0.196 |
| 02 | -0.634 | -2.56 | 1.719 | -2.56 | -0.466 | -4.38 | -0.322 | -1.431 | -0.836 | -4.38 | 0.944 | -1.952 | -0.575 | -2.56 | 1.448 | -2.56 |
| 03 | -2.21 | -4.38 | 0.687 | -3.103 | -4.38 | -4.38 | -2.21 | -3.103 | -1.952 | -1.431 | 2.516 | -0.998 | -2.56 | -2.21 | -1.431 | -2.21 |
| 04 | -3.103 | -4.38 | -1.431 | -3.103 | -3.103 | -3.103 | -1.576 | -2.56 | -1.431 | -1.746 | 2.646 | -1.576 | -4.38 | -2.21 | -0.914 | -3.103 |
| 05 | -3.103 | -1.952 | -3.103 | -2.21 | -4.38 | -4.38 | -4.38 | -1.191 | 0.428 | 1.47 | -2.21 | 2.191 | -2.56 | -2.21 | -2.56 | -2.21 |
| 06 | -1.746 | -4.38 | 0.363 | -3.103 | -3.103 | -1.952 | 1.409 | -1.09 | -4.38 | -3.103 | -1.952 | -3.103 | -1.746 | -2.21 | 2.199 | -1.746 |
| 07 | -3.103 | -4.38 | -1.952 | -1.576 | -3.103 | -4.38 | -3.103 | -1.576 | -0.196 | -1.431 | 1.742 | 2.068 | -3.103 | -4.38 | -0.998 | -1.746 |
| 08 | -3.103 | -3.103 | -0.236 | -2.56 | -4.38 | -4.38 | -1.576 | -3.103 | -1.952 | -2.21 | 1.765 | -1.746 | -1.746 | -1.746 | 2.073 | -1.576 |
| 09 | -3.103 | -4.38 | -1.431 | -2.56 | -3.103 | -2.21 | -1.952 | -3.103 | -1.09 | -0.998 | 2.437 | 0.969 | -4.38 | -4.38 | -0.998 | -2.21 |
| 10 | -4.38 | -1.191 | -3.103 | -2.56 | -2.21 | -0.998 | -4.38 | -4.38 | 0.217 | 2.101 | 0.107 | 0.509 | -1.576 | 0.566 | -0.415 | -1.431 |
| 11 | -0.634 | -0.914 | -0.836 | -1.576 | 1.327 | 0.75 | 0.078 | 1.292 | -0.519 | -1.952 | -1.431 | -0.157 | -0.836 | -0.697 | -0.466 | -0.836 |
| 12 | -0.836 | -0.368 | 1.227 | -0.236 | 0.047 | -0.368 | -0.12 | -0.634 | -1.09 | -0.466 | 0.243 | -1.952 | -1.09 | -0.157 | 1.189 | -0.415 |
| 13 | -0.998 | -1.304 | 0.191 | -1.191 | 0.047 | -0.914 | -0.998 | 0.047 | 0.509 | 0.217 | 1.401 | 0.637 | -1.576 | -0.764 | -0.157 | -0.519 |
| 14 | -0.415 | -1.431 | 0.703 | -0.998 | -0.519 | -1.09 | -0.157 | -0.519 | 0.489 | -0.466 | 1.318 | -0.697 | -0.836 | -0.575 | 0.879 | -0.836 |
| 15 | -0.016 | -0.12 | 0.107 | -1.09 | -0.236 | -0.575 | -2.21 | -1.191 | 0.566 | 0.931 | 1.128 | 0.489 | -1.191 | -0.575 | -0.12 | -1.952 |