Model info
| Transcription factor | MAFB (GeneCards) | ||||||||
| Model | MAFB_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 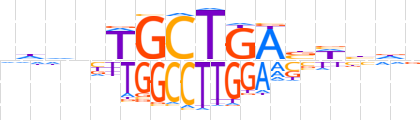 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dddnTGCTGAbWbdd | ||||||||
| Best auROC (human) | 0.684 | ||||||||
| Best auROC (mouse) | 0.969 | ||||||||
| Peak sets in benchmark (human) | 5 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 379 | ||||||||
| TF family | Maf-related factors {1.1.3} | ||||||||
| TF subfamily | Large Maf factors {1.1.3.1} | ||||||||
| HGNC | HGNC:6408 | ||||||||
| EntrezGene | GeneID:9935 (SSTAR profile) | ||||||||
| UniProt ID | MAFB_HUMAN | ||||||||
| UniProt AC | Q9Y5Q3 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MAFB expression | ||||||||
| ReMap ChIP-seq dataset list | MAFB datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 91.0 | 13.0 | 20.0 | 37.0 | 26.0 | 10.0 | 2.0 | 18.0 | 35.0 | 13.0 | 15.0 | 14.0 | 22.0 | 9.0 | 22.0 | 32.0 |
| 02 | 90.0 | 19.0 | 30.0 | 35.0 | 16.0 | 8.0 | 9.0 | 12.0 | 18.0 | 12.0 | 19.0 | 10.0 | 12.0 | 15.0 | 12.0 | 62.0 |
| 03 | 26.0 | 46.0 | 44.0 | 20.0 | 12.0 | 15.0 | 2.0 | 25.0 | 18.0 | 14.0 | 16.0 | 22.0 | 15.0 | 38.0 | 20.0 | 46.0 |
| 04 | 14.0 | 5.0 | 4.0 | 48.0 | 9.0 | 1.0 | 0.0 | 103.0 | 4.0 | 2.0 | 4.0 | 72.0 | 8.0 | 3.0 | 9.0 | 93.0 |
| 05 | 0.0 | 0.0 | 35.0 | 0.0 | 0.0 | 1.0 | 10.0 | 0.0 | 0.0 | 2.0 | 15.0 | 0.0 | 1.0 | 16.0 | 291.0 | 8.0 |
| 06 | 0.0 | 1.0 | 0.0 | 0.0 | 5.0 | 14.0 | 0.0 | 0.0 | 18.0 | 327.0 | 1.0 | 5.0 | 0.0 | 8.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 23.0 | 12.0 | 5.0 | 0.0 | 333.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 5.0 |
| 08 | 2.0 | 0.0 | 9.0 | 1.0 | 2.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 13.0 | 4.0 | 313.0 | 32.0 |
| 09 | 8.0 | 2.0 | 4.0 | 3.0 | 1.0 | 0.0 | 1.0 | 2.0 | 287.0 | 9.0 | 7.0 | 20.0 | 26.0 | 2.0 | 5.0 | 2.0 |
| 10 | 24.0 | 154.0 | 101.0 | 43.0 | 4.0 | 5.0 | 1.0 | 3.0 | 0.0 | 10.0 | 6.0 | 1.0 | 0.0 | 7.0 | 13.0 | 7.0 |
| 11 | 4.0 | 4.0 | 2.0 | 18.0 | 47.0 | 16.0 | 2.0 | 111.0 | 12.0 | 28.0 | 10.0 | 71.0 | 5.0 | 3.0 | 18.0 | 28.0 |
| 12 | 9.0 | 31.0 | 16.0 | 12.0 | 15.0 | 18.0 | 0.0 | 18.0 | 6.0 | 8.0 | 3.0 | 15.0 | 25.0 | 119.0 | 41.0 | 43.0 |
| 13 | 25.0 | 5.0 | 19.0 | 6.0 | 130.0 | 13.0 | 10.0 | 23.0 | 10.0 | 11.0 | 11.0 | 28.0 | 21.0 | 21.0 | 25.0 | 21.0 |
| 14 | 23.0 | 14.0 | 108.0 | 41.0 | 29.0 | 6.0 | 4.0 | 11.0 | 16.0 | 12.0 | 22.0 | 15.0 | 17.0 | 7.0 | 33.0 | 21.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.334 | -0.587 | -0.166 | 0.44 | 0.092 | -0.841 | -2.317 | -0.27 | 0.385 | -0.587 | -0.448 | -0.515 | -0.073 | -0.943 | -0.073 | 0.297 |
| 02 | 1.323 | -0.217 | 0.233 | 0.385 | -0.385 | -1.056 | -0.943 | -0.665 | -0.27 | -0.665 | -0.217 | -0.841 | -0.665 | -0.448 | -0.665 | 0.953 |
| 03 | 0.092 | 0.656 | 0.612 | -0.166 | -0.665 | -0.448 | -2.317 | 0.053 | -0.27 | -0.515 | -0.385 | -0.073 | -0.448 | 0.467 | -0.166 | 0.656 |
| 04 | -0.515 | -1.499 | -1.705 | 0.698 | -0.943 | -2.865 | -4.172 | 1.458 | -1.705 | -2.317 | -1.705 | 1.101 | -1.056 | -1.965 | -0.943 | 1.356 |
| 05 | -4.172 | -4.172 | 0.385 | -4.172 | -4.172 | -2.865 | -0.841 | -4.172 | -4.172 | -2.317 | -0.448 | -4.172 | -2.865 | -0.385 | 2.494 | -1.056 |
| 06 | -4.172 | -2.865 | -4.172 | -4.172 | -1.499 | -0.515 | -4.172 | -4.172 | -0.27 | 2.611 | -2.865 | -1.499 | -4.172 | -1.056 | -4.172 | -4.172 |
| 07 | -4.172 | -4.172 | -4.172 | -0.029 | -0.665 | -1.499 | -4.172 | 2.629 | -4.172 | -4.172 | -4.172 | -2.865 | -4.172 | -4.172 | -4.172 | -1.499 |
| 08 | -2.317 | -4.172 | -0.943 | -2.865 | -2.317 | -4.172 | -2.865 | -2.317 | -4.172 | -4.172 | -4.172 | -4.172 | -0.587 | -1.705 | 2.567 | 0.297 |
| 09 | -1.056 | -2.317 | -1.705 | -1.965 | -2.865 | -4.172 | -2.865 | -2.317 | 2.48 | -0.943 | -1.183 | -0.166 | 0.092 | -2.317 | -1.499 | -2.317 |
| 10 | 0.013 | 1.859 | 1.438 | 0.589 | -1.705 | -1.499 | -2.865 | -1.965 | -4.172 | -0.841 | -1.329 | -2.865 | -4.172 | -1.183 | -0.587 | -1.183 |
| 11 | -1.705 | -1.705 | -2.317 | -0.27 | 0.678 | -0.385 | -2.317 | 1.532 | -0.665 | 0.165 | -0.841 | 1.087 | -1.499 | -1.965 | -0.27 | 0.165 |
| 12 | -0.943 | 0.265 | -0.385 | -0.665 | -0.448 | -0.27 | -4.172 | -0.27 | -1.329 | -1.056 | -1.965 | -0.448 | 0.053 | 1.602 | 0.542 | 0.589 |
| 13 | 0.053 | -1.499 | -0.217 | -1.329 | 1.69 | -0.587 | -0.841 | -0.029 | -0.841 | -0.749 | -0.749 | 0.165 | -0.118 | -0.118 | 0.053 | -0.118 |
| 14 | -0.029 | -0.515 | 1.505 | 0.542 | 0.2 | -1.329 | -1.705 | -0.749 | -0.385 | -0.665 | -0.073 | -0.448 | -0.326 | -1.183 | 0.327 | -0.118 |