Model info
| Transcription factor | MAFK (GeneCards) | ||||||||
| Model | MAFK_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 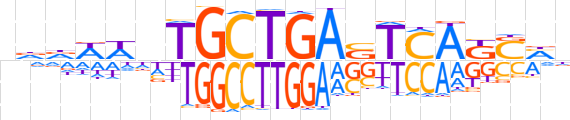 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nddWWnTGCTGASTCAKSMn | ||||||||
| Best auROC (human) | 0.994 | ||||||||
| Best auROC (mouse) | 0.993 | ||||||||
| Peak sets in benchmark (human) | 12 | ||||||||
| Peak sets in benchmark (mouse) | 11 | ||||||||
| Aligned words | 437 | ||||||||
| TF family | Maf-related factors {1.1.3} | ||||||||
| TF subfamily | Small Maf factors {1.1.3.2} | ||||||||
| HGNC | HGNC:6782 | ||||||||
| EntrezGene | GeneID:7975 (SSTAR profile) | ||||||||
| UniProt ID | MAFK_HUMAN | ||||||||
| UniProt AC | O60675 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MAFK expression | ||||||||
| ReMap ChIP-seq dataset list | MAFK datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 67.0 | 10.0 | 49.0 | 17.0 | 60.0 | 18.0 | 3.0 | 15.0 | 50.0 | 10.0 | 24.0 | 10.0 | 38.0 | 11.0 | 38.0 | 17.0 |
| 02 | 122.0 | 6.0 | 48.0 | 39.0 | 32.0 | 6.0 | 0.0 | 11.0 | 56.0 | 9.0 | 39.0 | 10.0 | 20.0 | 6.0 | 9.0 | 24.0 |
| 03 | 160.0 | 7.0 | 11.0 | 52.0 | 12.0 | 4.0 | 1.0 | 10.0 | 56.0 | 5.0 | 12.0 | 23.0 | 35.0 | 3.0 | 2.0 | 44.0 |
| 04 | 168.0 | 11.0 | 10.0 | 74.0 | 6.0 | 3.0 | 0.0 | 10.0 | 16.0 | 3.0 | 1.0 | 6.0 | 35.0 | 5.0 | 1.0 | 88.0 |
| 05 | 93.0 | 48.0 | 38.0 | 46.0 | 6.0 | 9.0 | 0.0 | 7.0 | 4.0 | 2.0 | 1.0 | 5.0 | 25.0 | 31.0 | 28.0 | 94.0 |
| 06 | 6.0 | 7.0 | 1.0 | 114.0 | 9.0 | 4.0 | 0.0 | 77.0 | 4.0 | 3.0 | 1.0 | 59.0 | 7.0 | 8.0 | 1.0 | 136.0 |
| 07 | 0.0 | 4.0 | 21.0 | 1.0 | 1.0 | 1.0 | 19.0 | 1.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 3.0 | 382.0 | 1.0 |
| 08 | 1.0 | 0.0 | 0.0 | 0.0 | 4.0 | 5.0 | 0.0 | 0.0 | 23.0 | 399.0 | 0.0 | 2.0 | 2.0 | 1.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 30.0 | 11.0 | 11.0 | 0.0 | 383.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 10 | 1.0 | 1.0 | 7.0 | 2.0 | 7.0 | 0.0 | 3.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 399.0 | 10.0 |
| 11 | 12.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 391.0 | 1.0 | 0.0 | 17.0 | 13.0 | 0.0 | 0.0 | 0.0 |
| 12 | 12.0 | 170.0 | 213.0 | 22.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 2.0 | 14.0 | 0.0 |
| 13 | 1.0 | 0.0 | 0.0 | 14.0 | 26.0 | 6.0 | 7.0 | 134.0 | 2.0 | 5.0 | 2.0 | 218.0 | 0.0 | 0.0 | 4.0 | 18.0 |
| 14 | 6.0 | 12.0 | 4.0 | 7.0 | 3.0 | 3.0 | 0.0 | 5.0 | 1.0 | 2.0 | 1.0 | 9.0 | 29.0 | 342.0 | 5.0 | 8.0 |
| 15 | 23.0 | 0.0 | 12.0 | 4.0 | 346.0 | 1.0 | 3.0 | 9.0 | 3.0 | 0.0 | 2.0 | 5.0 | 6.0 | 1.0 | 10.0 | 12.0 |
| 16 | 6.0 | 22.0 | 225.0 | 125.0 | 1.0 | 0.0 | 1.0 | 0.0 | 6.0 | 0.0 | 19.0 | 2.0 | 1.0 | 0.0 | 29.0 | 0.0 |
| 17 | 0.0 | 11.0 | 2.0 | 1.0 | 6.0 | 9.0 | 3.0 | 4.0 | 4.0 | 240.0 | 20.0 | 10.0 | 9.0 | 64.0 | 39.0 | 15.0 |
| 18 | 2.0 | 6.0 | 7.0 | 4.0 | 243.0 | 23.0 | 17.0 | 41.0 | 19.0 | 26.0 | 11.0 | 8.0 | 5.0 | 9.0 | 9.0 | 7.0 |
| 19 | 78.0 | 50.0 | 53.0 | 88.0 | 30.0 | 11.0 | 1.0 | 22.0 | 12.0 | 3.0 | 11.0 | 18.0 | 15.0 | 14.0 | 13.0 | 18.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.889 | -0.981 | 0.578 | -0.466 | 0.779 | -0.41 | -2.103 | -0.588 | 0.598 | -0.981 | -0.127 | -0.981 | 0.326 | -0.889 | 0.326 | -0.466 |
| 02 | 1.486 | -1.468 | 0.558 | 0.352 | 0.156 | -1.468 | -4.289 | -0.889 | 0.711 | -1.083 | 0.352 | -0.981 | -0.307 | -1.468 | -1.083 | -0.127 |
| 03 | 1.756 | -1.322 | -0.889 | 0.637 | -0.805 | -1.844 | -2.999 | -0.981 | 0.711 | -1.638 | -0.805 | -0.169 | 0.245 | -2.103 | -2.454 | 0.472 |
| 04 | 1.805 | -0.889 | -0.981 | 0.988 | -1.468 | -2.103 | -4.289 | -0.981 | -0.525 | -2.103 | -2.999 | -1.468 | 0.245 | -1.638 | -2.999 | 1.16 |
| 05 | 1.216 | 0.558 | 0.326 | 0.516 | -1.468 | -1.083 | -4.289 | -1.322 | -1.844 | -2.454 | -2.999 | -1.638 | -0.087 | 0.125 | 0.025 | 1.226 |
| 06 | -1.468 | -1.322 | -2.999 | 1.418 | -1.083 | -1.844 | -4.289 | 1.028 | -1.844 | -2.103 | -2.999 | 0.763 | -1.322 | -1.195 | -2.999 | 1.594 |
| 07 | -4.289 | -1.844 | -0.259 | -2.999 | -2.999 | -2.999 | -0.357 | -2.999 | -4.289 | -2.999 | -2.454 | -4.289 | -4.289 | -2.103 | 2.625 | -2.999 |
| 08 | -2.999 | -4.289 | -4.289 | -4.289 | -1.844 | -1.638 | -4.289 | -4.289 | -0.169 | 2.669 | -4.289 | -2.454 | -2.454 | -2.999 | -4.289 | -4.289 |
| 09 | -4.289 | -4.289 | -4.289 | 0.093 | -0.889 | -0.889 | -4.289 | 2.628 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -4.289 | -2.454 |
| 10 | -2.999 | -2.999 | -1.322 | -2.454 | -1.322 | -4.289 | -2.103 | -2.999 | -4.289 | -4.289 | -4.289 | -4.289 | -1.468 | -4.289 | 2.669 | -0.981 |
| 11 | -0.805 | -4.289 | -2.999 | -2.999 | -2.999 | -4.289 | -4.289 | -4.289 | 2.649 | -2.999 | -4.289 | -0.466 | -0.727 | -4.289 | -4.289 | -4.289 |
| 12 | -0.805 | 1.817 | 2.042 | -0.213 | -2.999 | -4.289 | -4.289 | -4.289 | -4.289 | -2.999 | -4.289 | -4.289 | -2.454 | -2.454 | -0.655 | -4.289 |
| 13 | -2.999 | -4.289 | -4.289 | -0.655 | -0.049 | -1.468 | -1.322 | 1.58 | -2.454 | -1.638 | -2.454 | 2.065 | -4.289 | -4.289 | -1.844 | -0.41 |
| 14 | -1.468 | -0.805 | -1.844 | -1.322 | -2.103 | -2.103 | -4.289 | -1.638 | -2.999 | -2.454 | -2.999 | -1.083 | 0.059 | 2.515 | -1.638 | -1.195 |
| 15 | -0.169 | -4.289 | -0.805 | -1.844 | 2.526 | -2.999 | -2.103 | -1.083 | -2.103 | -4.289 | -2.454 | -1.638 | -1.468 | -2.999 | -0.981 | -0.805 |
| 16 | -1.468 | -0.213 | 2.097 | 1.51 | -2.999 | -4.289 | -2.999 | -4.289 | -1.468 | -4.289 | -0.357 | -2.454 | -2.999 | -4.289 | 0.059 | -4.289 |
| 17 | -4.289 | -0.889 | -2.454 | -2.999 | -1.468 | -1.083 | -2.103 | -1.844 | -1.844 | 2.161 | -0.307 | -0.981 | -1.083 | 0.844 | 0.352 | -0.588 |
| 18 | -2.454 | -1.468 | -1.322 | -1.844 | 2.173 | -0.169 | -0.466 | 0.402 | -0.357 | -0.049 | -0.889 | -1.195 | -1.638 | -1.083 | -1.083 | -1.322 |
| 19 | 1.04 | 0.598 | 0.656 | 1.16 | 0.093 | -0.889 | -2.999 | -0.213 | -0.805 | -2.103 | -0.889 | -0.41 | -0.588 | -0.655 | -0.727 | -0.41 |