Model info
| Transcription factor | MEF2B (GeneCards) | ||||||||
| Model | MEF2B_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 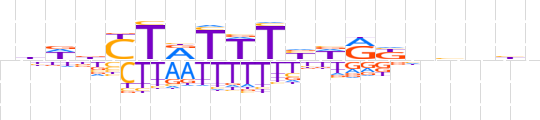 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbWbYTRTWTbWRbnhnhn | ||||||||
| Best auROC (human) | 0.962 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 23 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 373 | ||||||||
| TF family | Regulators of differentiation {5.1.1} | ||||||||
| TF subfamily | MEF-2 {5.1.1.1} | ||||||||
| HGNC | HGNC:6995 | ||||||||
| EntrezGene | GeneID:100271849; 4207 (SSTAR profile) | ||||||||
| UniProt ID | MEF2B_HUMAN | ||||||||
| UniProt AC | Q02080 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MEF2B expression | ||||||||
| ReMap ChIP-seq dataset list | MEF2B datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 16.0 | 9.0 | 26.0 | 28.0 | 14.0 | 20.0 | 3.0 | 21.0 | 22.0 | 31.0 | 16.0 | 70.0 | 9.0 | 14.0 | 26.0 | 40.0 |
| 02 | 17.0 | 1.0 | 19.0 | 24.0 | 14.0 | 4.0 | 6.0 | 50.0 | 28.0 | 2.0 | 11.0 | 30.0 | 23.0 | 3.0 | 41.0 | 92.0 |
| 03 | 13.0 | 8.0 | 39.0 | 22.0 | 5.0 | 2.0 | 1.0 | 2.0 | 8.0 | 14.0 | 23.0 | 32.0 | 14.0 | 43.0 | 49.0 | 90.0 |
| 04 | 0.0 | 34.0 | 0.0 | 6.0 | 3.0 | 53.0 | 2.0 | 9.0 | 3.0 | 68.0 | 7.0 | 34.0 | 2.0 | 116.0 | 11.0 | 17.0 |
| 05 | 2.0 | 1.0 | 0.0 | 5.0 | 3.0 | 34.0 | 1.0 | 233.0 | 1.0 | 1.0 | 0.0 | 18.0 | 2.0 | 4.0 | 1.0 | 59.0 |
| 06 | 6.0 | 0.0 | 2.0 | 0.0 | 21.0 | 2.0 | 5.0 | 12.0 | 1.0 | 0.0 | 1.0 | 0.0 | 212.0 | 26.0 | 59.0 | 18.0 |
| 07 | 17.0 | 5.0 | 1.0 | 217.0 | 6.0 | 10.0 | 0.0 | 12.0 | 2.0 | 13.0 | 1.0 | 51.0 | 1.0 | 6.0 | 0.0 | 23.0 |
| 08 | 9.0 | 0.0 | 3.0 | 14.0 | 3.0 | 5.0 | 6.0 | 20.0 | 0.0 | 0.0 | 0.0 | 2.0 | 23.0 | 13.0 | 24.0 | 243.0 |
| 09 | 1.0 | 3.0 | 0.0 | 31.0 | 0.0 | 1.0 | 0.0 | 17.0 | 3.0 | 4.0 | 4.0 | 22.0 | 9.0 | 24.0 | 0.0 | 246.0 |
| 10 | 4.0 | 0.0 | 1.0 | 8.0 | 2.0 | 4.0 | 1.0 | 25.0 | 1.0 | 0.0 | 0.0 | 3.0 | 37.0 | 63.0 | 47.0 | 169.0 |
| 11 | 13.0 | 1.0 | 7.0 | 23.0 | 17.0 | 5.0 | 1.0 | 44.0 | 4.0 | 7.0 | 7.0 | 31.0 | 25.0 | 26.0 | 31.0 | 123.0 |
| 12 | 15.0 | 0.0 | 43.0 | 1.0 | 24.0 | 2.0 | 5.0 | 8.0 | 8.0 | 0.0 | 37.0 | 1.0 | 78.0 | 0.0 | 131.0 | 12.0 |
| 13 | 12.0 | 14.0 | 74.0 | 25.0 | 0.0 | 1.0 | 1.0 | 0.0 | 19.0 | 18.0 | 121.0 | 58.0 | 1.0 | 4.0 | 12.0 | 5.0 |
| 14 | 8.0 | 14.0 | 3.0 | 7.0 | 13.0 | 11.0 | 0.0 | 13.0 | 67.0 | 68.0 | 25.0 | 48.0 | 23.0 | 19.0 | 25.0 | 21.0 |
| 15 | 36.0 | 35.0 | 17.0 | 23.0 | 43.0 | 32.0 | 2.0 | 35.0 | 7.0 | 22.0 | 5.0 | 19.0 | 20.0 | 24.0 | 12.0 | 33.0 |
| 16 | 30.0 | 30.0 | 29.0 | 17.0 | 45.0 | 30.0 | 5.0 | 33.0 | 9.0 | 15.0 | 5.0 | 7.0 | 16.0 | 37.0 | 20.0 | 37.0 |
| 17 | 13.0 | 34.0 | 17.0 | 36.0 | 26.0 | 28.0 | 3.0 | 55.0 | 8.0 | 19.0 | 6.0 | 26.0 | 8.0 | 23.0 | 27.0 | 36.0 |
| 18 | 18.0 | 4.0 | 24.0 | 9.0 | 33.0 | 15.0 | 3.0 | 53.0 | 12.0 | 11.0 | 19.0 | 11.0 | 25.0 | 22.0 | 56.0 | 50.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.348 | -0.906 | 0.129 | 0.202 | -0.478 | -0.129 | -1.929 | -0.081 | -0.036 | 0.302 | -0.348 | 1.11 | -0.906 | -0.478 | 0.129 | 0.555 |
| 02 | -0.289 | -2.829 | -0.18 | 0.05 | -0.478 | -1.669 | -1.292 | 0.776 | 0.202 | -2.281 | -0.712 | 0.27 | 0.008 | -1.929 | 0.579 | 1.382 |
| 03 | -0.55 | -1.019 | 0.53 | -0.036 | -1.463 | -2.281 | -2.829 | -2.281 | -1.019 | -0.478 | 0.008 | 0.334 | -0.478 | 0.626 | 0.756 | 1.361 |
| 04 | -4.141 | 0.394 | -4.141 | -1.292 | -1.929 | 0.834 | -2.281 | -0.906 | -1.929 | 1.082 | -1.146 | 0.394 | -2.281 | 1.613 | -0.712 | -0.289 |
| 05 | -2.281 | -2.829 | -4.141 | -1.463 | -1.929 | 0.394 | -2.829 | 2.309 | -2.829 | -2.829 | -4.141 | -0.233 | -2.281 | -1.669 | -2.829 | 0.94 |
| 06 | -1.292 | -4.141 | -2.281 | -4.141 | -0.081 | -2.281 | -1.463 | -0.628 | -2.829 | -4.141 | -2.829 | -4.141 | 2.215 | 0.129 | 0.94 | -0.233 |
| 07 | -0.289 | -1.463 | -2.829 | 2.238 | -1.292 | -0.805 | -4.141 | -0.628 | -2.281 | -0.55 | -2.829 | 0.796 | -2.829 | -1.292 | -4.141 | 0.008 |
| 08 | -0.906 | -4.141 | -1.929 | -0.478 | -1.929 | -1.463 | -1.292 | -0.129 | -4.141 | -4.141 | -4.141 | -2.281 | 0.008 | -0.55 | 0.05 | 2.351 |
| 09 | -2.829 | -1.929 | -4.141 | 0.302 | -4.141 | -2.829 | -4.141 | -0.289 | -1.929 | -1.669 | -1.669 | -0.036 | -0.906 | 0.05 | -4.141 | 2.363 |
| 10 | -1.669 | -4.141 | -2.829 | -1.019 | -2.281 | -1.669 | -2.829 | 0.09 | -2.829 | -4.141 | -4.141 | -1.929 | 0.477 | 1.006 | 0.715 | 1.989 |
| 11 | -0.55 | -2.829 | -1.146 | 0.008 | -0.289 | -1.463 | -2.829 | 0.649 | -1.669 | -1.146 | -1.146 | 0.302 | 0.09 | 0.129 | 0.302 | 1.672 |
| 12 | -0.411 | -4.141 | 0.626 | -2.829 | 0.05 | -2.281 | -1.463 | -1.019 | -1.019 | -4.141 | 0.477 | -2.829 | 1.218 | -4.141 | 1.735 | -0.628 |
| 13 | -0.628 | -0.478 | 1.166 | 0.09 | -4.141 | -2.829 | -2.829 | -4.141 | -0.18 | -0.233 | 1.655 | 0.923 | -2.829 | -1.669 | -0.628 | -1.463 |
| 14 | -1.019 | -0.478 | -1.929 | -1.146 | -0.55 | -0.712 | -4.141 | -0.55 | 1.067 | 1.082 | 0.09 | 0.736 | 0.008 | -0.18 | 0.09 | -0.081 |
| 15 | 0.45 | 0.422 | -0.289 | 0.008 | 0.626 | 0.334 | -2.281 | 0.422 | -1.146 | -0.036 | -1.463 | -0.18 | -0.129 | 0.05 | -0.628 | 0.364 |
| 16 | 0.27 | 0.27 | 0.237 | -0.289 | 0.671 | 0.27 | -1.463 | 0.364 | -0.906 | -0.411 | -1.463 | -1.146 | -0.348 | 0.477 | -0.129 | 0.477 |
| 17 | -0.55 | 0.394 | -0.289 | 0.45 | 0.129 | 0.202 | -1.929 | 0.871 | -1.019 | -0.18 | -1.292 | 0.129 | -1.019 | 0.008 | 0.166 | 0.45 |
| 18 | -0.233 | -1.669 | 0.05 | -0.906 | 0.364 | -0.411 | -1.929 | 0.834 | -0.628 | -0.712 | -0.18 | -0.712 | 0.09 | -0.036 | 0.889 | 0.776 |