Model info
| Transcription factor | MYCN (GeneCards) | ||||||||
| Model | MYCN_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 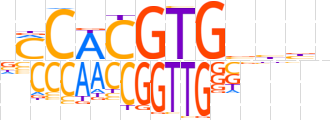 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvvvCACGTGKv | ||||||||
| Best auROC (human) | 0.935 | ||||||||
| Best auROC (mouse) | 0.899 | ||||||||
| Peak sets in benchmark (human) | 32 | ||||||||
| Peak sets in benchmark (mouse) | 10 | ||||||||
| Aligned words | 402 | ||||||||
| TF family | bHLH-ZIP factors {1.2.6} | ||||||||
| TF subfamily | Myc / Max factors {1.2.6.5} | ||||||||
| HGNC | HGNC:7559 | ||||||||
| EntrezGene | GeneID:4613 (SSTAR profile) | ||||||||
| UniProt ID | MYCN_HUMAN | ||||||||
| UniProt AC | P04198 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | MYCN expression | ||||||||
| ReMap ChIP-seq dataset list | MYCN datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 23.0 | 4.0 | 35.0 | 2.0 | 36.0 | 32.0 | 42.0 | 14.0 | 38.0 | 32.0 | 61.0 | 5.0 | 13.0 | 10.0 | 38.0 | 6.0 |
| 02 | 41.0 | 18.0 | 43.0 | 8.0 | 34.0 | 12.0 | 22.0 | 10.0 | 64.0 | 48.0 | 51.0 | 13.0 | 7.0 | 4.0 | 11.0 | 5.0 |
| 03 | 33.0 | 40.0 | 68.0 | 5.0 | 16.0 | 14.0 | 50.0 | 2.0 | 14.0 | 64.0 | 16.0 | 33.0 | 9.0 | 7.0 | 16.0 | 4.0 |
| 04 | 1.0 | 71.0 | 0.0 | 0.0 | 0.0 | 125.0 | 0.0 | 0.0 | 0.0 | 149.0 | 0.0 | 1.0 | 0.0 | 44.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 1.0 | 0.0 | 376.0 | 1.0 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 06 | 0.0 | 366.0 | 6.0 | 4.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 14.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 39.0 | 2.0 | 340.0 | 0.0 | 3.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 |
| 08 | 1.0 | 0.0 | 0.0 | 41.0 | 2.0 | 0.0 | 0.0 | 0.0 | 39.0 | 28.0 | 5.0 | 275.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 42.0 | 0.0 | 0.0 | 3.0 | 23.0 | 2.0 | 2.0 | 0.0 | 3.0 | 0.0 | 1.0 | 0.0 | 308.0 | 7.0 |
| 10 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 2.0 | 1.0 | 21.0 | 28.0 | 293.0 | 34.0 | 1.0 | 1.0 | 7.0 | 0.0 |
| 11 | 3.0 | 10.0 | 5.0 | 4.0 | 7.0 | 10.0 | 9.0 | 3.0 | 51.0 | 121.0 | 87.0 | 44.0 | 7.0 | 15.0 | 12.0 | 3.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.06 | -1.736 | 0.355 | -2.347 | 0.383 | 0.266 | 0.535 | -0.546 | 0.436 | 0.266 | 0.906 | -1.53 | -0.618 | -0.872 | 0.436 | -1.359 |
| 02 | 0.511 | -0.3 | 0.559 | -1.086 | 0.326 | -0.696 | -0.103 | -0.872 | 0.953 | 0.668 | 0.728 | -0.618 | -1.213 | -1.736 | -0.78 | -1.53 |
| 03 | 0.296 | 0.487 | 1.014 | -1.53 | -0.416 | -0.546 | 0.708 | -2.347 | -0.546 | 0.953 | -0.416 | 0.296 | -0.973 | -1.213 | -0.416 | -1.736 |
| 04 | -2.894 | 1.057 | -4.197 | -4.197 | -4.197 | 1.62 | -4.197 | -4.197 | -4.197 | 1.795 | -4.197 | -2.894 | -4.197 | 0.581 | -4.197 | -4.197 |
| 05 | -4.197 | -4.197 | -2.894 | -4.197 | 2.719 | -2.894 | -0.696 | -4.197 | -4.197 | -4.197 | -4.197 | -4.197 | -4.197 | -4.197 | -2.894 | -4.197 |
| 06 | -4.197 | 2.692 | -1.359 | -1.736 | -4.197 | -2.894 | -4.197 | -4.197 | -4.197 | -0.546 | -4.197 | -4.197 | -4.197 | -4.197 | -4.197 | -4.197 |
| 07 | -4.197 | -4.197 | -4.197 | -4.197 | 0.462 | -2.347 | 2.619 | -4.197 | -1.995 | -4.197 | -1.995 | -4.197 | -4.197 | -4.197 | -1.736 | -4.197 |
| 08 | -2.894 | -4.197 | -4.197 | 0.511 | -2.347 | -4.197 | -4.197 | -4.197 | 0.462 | 0.134 | -1.53 | 2.407 | -4.197 | -4.197 | -4.197 | -4.197 |
| 09 | -4.197 | -4.197 | 0.535 | -4.197 | -4.197 | -1.995 | -0.06 | -2.347 | -2.347 | -4.197 | -1.995 | -4.197 | -2.894 | -4.197 | 2.52 | -1.213 |
| 10 | -4.197 | -4.197 | -2.894 | -2.347 | -4.197 | -4.197 | -2.347 | -2.894 | -0.149 | 0.134 | 2.47 | 0.326 | -2.894 | -2.894 | -1.213 | -4.197 |
| 11 | -1.995 | -0.872 | -1.53 | -1.736 | -1.213 | -0.872 | -0.973 | -1.995 | 0.728 | 1.588 | 1.259 | 0.581 | -1.213 | -0.479 | -0.696 | -1.995 |