Model info
| Transcription factor | SMAD4 (GeneCards) | ||||||||
| Model | SMAD4_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 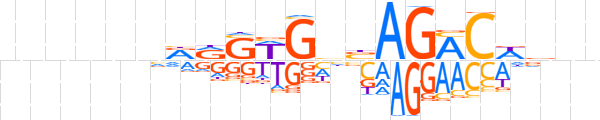 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnbWGKCTvdCWSYYnnnnnn | ||||||||
| Best auROC (human) | 0.882 | ||||||||
| Best auROC (mouse) | 0.926 | ||||||||
| Peak sets in benchmark (human) | 24 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | SMAD factors {7.1.1} | ||||||||
| TF subfamily | Co-activating Smads (Co-Smad) {7.1.1.2} | ||||||||
| HGNC | HGNC:6770 | ||||||||
| EntrezGene | GeneID:4089 (SSTAR profile) | ||||||||
| UniProt ID | SMAD4_HUMAN | ||||||||
| UniProt AC | Q13485 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SMAD4 expression | ||||||||
| ReMap ChIP-seq dataset list | SMAD4 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 23.446 | 22.009 | 30.427 | 26.517 | 39.376 | 65.675 | 9.032 | 64.517 | 18.436 | 30.07 | 24.048 | 18.369 | 8.754 | 31.459 | 48.936 | 38.929 |
| 02 | 14.483 | 28.187 | 35.26 | 12.081 | 31.954 | 66.535 | 22.353 | 28.371 | 13.513 | 47.011 | 38.883 | 13.036 | 14.131 | 58.009 | 43.752 | 32.441 |
| 03 | 25.506 | 6.1 | 7.123 | 35.352 | 60.885 | 13.551 | 2.373 | 122.933 | 28.173 | 29.578 | 13.328 | 69.169 | 17.057 | 3.284 | 7.508 | 58.081 |
| 04 | 0.0 | 0.857 | 129.097 | 1.667 | 0.868 | 2.792 | 44.142 | 4.711 | 0.0 | 2.101 | 27.14 | 1.09 | 3.973 | 7.551 | 271.339 | 2.671 |
| 05 | 0.0 | 0.0 | 0.0 | 4.841 | 0.0 | 2.012 | 1.038 | 10.251 | 8.429 | 20.788 | 79.513 | 362.987 | 0.0 | 3.703 | 1.955 | 4.482 |
| 06 | 0.868 | 7.561 | 0.0 | 0.0 | 0.899 | 23.682 | 0.0 | 1.923 | 0.966 | 76.855 | 0.846 | 3.839 | 0.917 | 378.009 | 0.0 | 3.635 |
| 07 | 0.0 | 0.0 | 0.0 | 3.651 | 2.978 | 2.141 | 0.0 | 480.988 | 0.0 | 0.0 | 0.0 | 0.846 | 0.0 | 0.0 | 0.0 | 9.396 |
| 08 | 1.736 | 0.0 | 1.242 | 0.0 | 0.0 | 1.081 | 0.0 | 1.06 | 0.0 | 0.0 | 0.0 | 0.0 | 106.431 | 112.363 | 254.297 | 21.79 |
| 09 | 7.564 | 4.021 | 48.917 | 47.665 | 57.739 | 11.072 | 3.799 | 40.834 | 52.15 | 39.485 | 102.072 | 61.832 | 3.61 | 0.0 | 7.902 | 11.339 |
| 10 | 1.086 | 104.667 | 8.827 | 6.484 | 3.097 | 48.493 | 0.0 | 2.989 | 1.642 | 154.439 | 4.665 | 1.943 | 2.047 | 148.048 | 5.472 | 6.102 |
| 11 | 0.0 | 1.933 | 4.175 | 1.764 | 343.516 | 29.811 | 28.868 | 53.451 | 7.695 | 3.112 | 0.951 | 7.206 | 2.653 | 2.217 | 6.443 | 6.205 |
| 12 | 21.895 | 266.678 | 46.987 | 18.303 | 1.834 | 32.559 | 0.0 | 2.68 | 3.063 | 29.447 | 5.488 | 2.439 | 0.0 | 57.633 | 6.369 | 4.625 |
| 13 | 0.908 | 16.307 | 7.595 | 1.983 | 52.3 | 237.49 | 1.214 | 95.313 | 3.822 | 36.464 | 7.671 | 10.886 | 5.516 | 12.442 | 6.322 | 3.767 |
| 14 | 11.268 | 9.327 | 16.346 | 25.606 | 29.825 | 45.476 | 6.196 | 221.205 | 2.704 | 3.953 | 6.831 | 9.313 | 10.197 | 27.23 | 20.409 | 54.113 |
| 15 | 6.046 | 17.554 | 22.984 | 7.411 | 29.399 | 16.717 | 4.084 | 35.786 | 10.073 | 11.515 | 16.003 | 12.191 | 44.336 | 57.351 | 105.681 | 102.868 |
| 16 | 19.091 | 21.538 | 26.701 | 22.525 | 23.679 | 32.163 | 0.908 | 46.388 | 31.315 | 48.868 | 26.13 | 42.439 | 13.609 | 43.457 | 52.498 | 48.693 |
| 17 | 19.811 | 19.822 | 22.102 | 25.958 | 24.486 | 40.24 | 1.944 | 79.355 | 26.755 | 18.179 | 19.589 | 41.713 | 42.554 | 23.75 | 41.958 | 51.783 |
| 18 | 64.462 | 15.197 | 22.572 | 11.376 | 37.94 | 28.006 | 3.313 | 32.731 | 28.488 | 27.413 | 18.056 | 11.636 | 55.135 | 28.097 | 62.591 | 52.986 |
| 19 | 64.527 | 22.156 | 37.042 | 62.3 | 29.331 | 16.102 | 2.879 | 50.4 | 20.238 | 18.171 | 31.639 | 36.483 | 22.093 | 30.274 | 27.275 | 29.089 |
| 20 | 28.471 | 12.749 | 44.707 | 50.261 | 27.804 | 30.75 | 0.0 | 28.15 | 11.115 | 26.824 | 35.214 | 25.682 | 17.912 | 65.513 | 55.301 | 39.546 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.283 | -0.345 | -0.026 | -0.162 | 0.229 | 0.736 | -1.212 | 0.719 | -0.519 | -0.038 | -0.258 | -0.523 | -1.241 | 0.007 | 0.444 | 0.217 |
| 02 | -0.755 | -0.102 | 0.119 | -0.931 | 0.022 | 0.749 | -0.33 | -0.095 | -0.822 | 0.404 | 0.216 | -0.857 | -0.779 | 0.613 | 0.333 | 0.037 |
| 03 | -0.2 | -1.584 | -1.438 | 0.122 | 0.661 | -0.82 | -2.439 | 1.36 | -0.102 | -0.054 | -0.836 | 0.788 | -0.595 | -2.154 | -1.388 | 0.614 |
| 04 | -4.4 | -3.235 | 1.409 | -2.734 | -3.226 | -2.297 | 0.342 | -1.825 | -4.4 | -2.542 | -0.139 | -3.063 | -1.982 | -1.383 | 2.15 | -2.336 |
| 05 | -4.4 | -4.4 | -4.4 | -1.8 | -4.4 | -2.579 | -3.099 | -1.09 | -1.278 | -0.401 | 0.926 | 2.441 | -4.4 | -2.045 | -2.603 | -1.871 |
| 06 | -3.226 | -1.381 | -4.4 | -4.4 | -3.202 | -0.273 | -4.4 | -2.617 | -3.151 | 0.893 | -3.244 | -2.013 | -3.188 | 2.482 | -4.4 | -2.062 |
| 07 | -4.4 | -4.4 | -4.4 | -2.058 | -2.241 | -2.526 | -4.4 | 2.722 | -4.4 | -4.4 | -4.4 | -3.244 | -4.4 | -4.4 | -4.4 | -1.174 |
| 08 | -2.701 | -4.4 | -2.966 | -4.4 | -4.4 | -3.07 | -4.4 | -3.084 | -4.4 | -4.4 | -4.4 | -4.4 | 1.217 | 1.271 | 2.086 | -0.355 |
| 09 | -1.381 | -1.971 | 0.444 | 0.418 | 0.608 | -1.015 | -2.022 | 0.265 | 0.507 | 0.231 | 1.175 | 0.676 | -2.069 | -4.4 | -1.339 | -0.992 |
| 10 | -3.066 | 1.2 | -1.234 | -1.527 | -2.206 | 0.435 | -4.4 | -2.237 | -2.746 | 1.588 | -1.834 | -2.608 | -2.564 | 1.546 | -1.686 | -1.584 |
| 11 | -4.4 | -2.612 | -1.936 | -2.688 | 2.386 | -0.047 | -0.078 | 0.532 | -1.365 | -2.201 | -3.162 | -1.427 | -2.342 | -2.497 | -1.533 | -1.568 |
| 12 | -0.351 | 2.133 | 0.404 | -0.526 | -2.656 | 0.041 | -4.4 | -2.333 | -2.215 | -0.059 | -1.683 | -2.415 | -4.4 | 0.606 | -1.544 | -1.842 |
| 13 | -3.195 | -0.639 | -1.377 | -2.591 | 0.51 | 2.017 | -2.983 | 1.107 | -2.017 | 0.153 | -1.368 | -1.032 | -1.679 | -0.903 | -1.551 | -2.03 |
| 14 | -0.999 | -1.181 | -0.637 | -0.197 | -0.046 | 0.371 | -1.57 | 1.946 | -2.325 | -1.986 | -1.478 | -1.182 | -1.095 | -0.136 | -0.42 | 0.544 |
| 15 | -1.593 | -0.567 | -0.303 | -1.4 | -0.06 | -0.615 | -1.956 | 0.134 | -1.107 | -0.978 | -0.658 | -0.922 | 0.346 | 0.602 | 1.21 | 1.183 |
| 16 | -0.485 | -0.367 | -0.155 | -0.323 | -0.274 | 0.028 | -3.195 | 0.391 | 0.002 | 0.443 | -0.177 | 0.303 | -0.816 | 0.326 | 0.514 | 0.439 |
| 17 | -0.449 | -0.448 | -0.341 | -0.183 | -0.241 | 0.25 | -2.607 | 0.924 | -0.153 | -0.533 | -0.46 | 0.286 | 0.305 | -0.271 | 0.292 | 0.5 |
| 18 | 0.718 | -0.708 | -0.321 | -0.989 | 0.192 | -0.108 | -2.146 | 0.046 | -0.091 | -0.129 | -0.54 | -0.967 | 0.562 | -0.105 | 0.688 | 0.523 |
| 19 | 0.719 | -0.339 | 0.168 | 0.684 | -0.063 | -0.652 | -2.27 | 0.473 | -0.428 | -0.533 | 0.012 | 0.153 | -0.342 | -0.031 | -0.134 | -0.071 |
| 20 | -0.092 | -0.879 | 0.354 | 0.471 | -0.115 | -0.016 | -4.4 | -0.103 | -1.012 | -0.151 | 0.118 | -0.194 | -0.547 | 0.734 | 0.565 | 0.233 |