Model info
| Transcription factor | TAL1 (GeneCards) | ||||||||
| Model | TAL1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 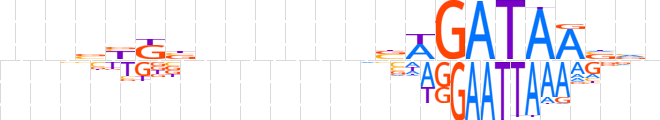 | ||||||||
| LOGO (reverse complement) | 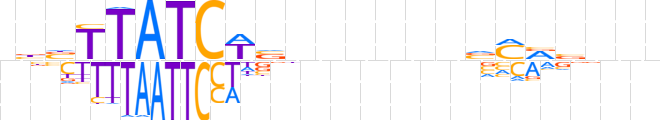 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnnbYKbnnnnnnbWGATAAvvn | ||||||||
| Best auROC (human) | 0.99 | ||||||||
| Best auROC (mouse) | 0.977 | ||||||||
| Peak sets in benchmark (human) | 68 | ||||||||
| Peak sets in benchmark (mouse) | 76 | ||||||||
| Aligned words | 503 | ||||||||
| TF family | Tal-related factors {1.2.3} | ||||||||
| TF subfamily | Tal / HEN-like factors {1.2.3.1} | ||||||||
| HGNC | HGNC:11556 | ||||||||
| EntrezGene | GeneID:6886 (SSTAR profile) | ||||||||
| UniProt ID | TAL1_HUMAN | ||||||||
| UniProt AC | P17542 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | TAL1 expression | ||||||||
| ReMap ChIP-seq dataset list | TAL1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 29.063 | 25.063 | 34.063 | 16.063 | 86.063 | 37.063 | 26.063 | 23.063 | 17.063 | 49.063 | 59.063 | 20.063 | 4.063 | 16.063 | 47.063 | 11.063 |
| 02 | 19.063 | 25.063 | 55.063 | 37.063 | 42.063 | 29.063 | 6.063 | 50.063 | 15.063 | 82.063 | 32.063 | 37.063 | 5.063 | 25.063 | 24.063 | 16.063 |
| 03 | 6.063 | 20.063 | 45.063 | 10.063 | 28.063 | 80.063 | 9.063 | 44.063 | 13.063 | 60.063 | 32.063 | 12.063 | 10.063 | 79.063 | 42.063 | 9.063 |
| 04 | 4.063 | 14.063 | 12.063 | 27.063 | 17.063 | 26.063 | 3.063 | 193.063 | 8.063 | 32.063 | 17.063 | 71.063 | 5.063 | 14.063 | 43.063 | 13.063 |
| 05 | 4.063 | 4.063 | 19.063 | 7.063 | 10.063 | 6.063 | 5.063 | 65.063 | 6.063 | 6.063 | 19.063 | 44.063 | 1.063 | 6.063 | 283.063 | 14.063 |
| 06 | 3.063 | 2.063 | 16.063 | 0.063 | 2.063 | 12.063 | 2.063 | 6.063 | 21.063 | 117.063 | 111.063 | 77.063 | 1.063 | 7.063 | 108.063 | 14.063 |
| 07 | 8.063 | 2.063 | 17.063 | 0.063 | 48.063 | 27.063 | 10.063 | 53.063 | 35.063 | 68.063 | 67.063 | 67.063 | 20.063 | 14.063 | 39.063 | 24.063 |
| 08 | 27.063 | 25.063 | 46.063 | 13.063 | 47.063 | 32.063 | 12.063 | 20.063 | 37.063 | 38.063 | 35.063 | 23.063 | 13.063 | 29.063 | 71.063 | 31.063 |
| 09 | 41.063 | 20.063 | 56.063 | 7.063 | 36.063 | 36.063 | 11.063 | 41.063 | 34.063 | 46.063 | 56.063 | 28.063 | 11.063 | 11.063 | 47.063 | 18.063 |
| 10 | 30.063 | 23.063 | 58.063 | 11.063 | 29.063 | 28.063 | 14.063 | 42.063 | 36.063 | 51.063 | 66.063 | 17.063 | 3.063 | 27.063 | 34.063 | 30.063 |
| 11 | 18.063 | 18.063 | 48.063 | 14.063 | 25.063 | 61.063 | 5.063 | 38.063 | 27.063 | 57.063 | 58.063 | 30.063 | 12.063 | 25.063 | 38.063 | 25.063 |
| 12 | 23.063 | 14.063 | 35.063 | 10.063 | 68.063 | 41.063 | 9.063 | 43.063 | 32.063 | 36.063 | 54.063 | 27.063 | 19.063 | 24.063 | 45.063 | 19.063 |
| 13 | 14.063 | 77.063 | 42.063 | 9.063 | 11.063 | 74.063 | 7.063 | 23.063 | 18.063 | 72.063 | 44.063 | 9.063 | 6.063 | 39.063 | 42.063 | 12.063 |
| 14 | 38.063 | 3.063 | 2.063 | 6.063 | 141.063 | 4.063 | 0.063 | 117.063 | 91.063 | 3.063 | 1.063 | 40.063 | 27.063 | 1.063 | 0.063 | 25.063 |
| 15 | 0.063 | 0.063 | 297.063 | 0.063 | 0.063 | 0.063 | 11.063 | 0.063 | 0.063 | 0.063 | 3.063 | 0.063 | 0.063 | 0.063 | 188.063 | 0.063 |
| 16 | 0.25 | 0.0 | 0.0 | 0.0 | 0.25 | 0.0 | 0.0 | 0.0 | 496.25 | 1.0 | 0.0 | 2.0 | 0.25 | 0.0 | 0.0 | 0.0 |
| 17 | 0.0 | 2.0 | 0.0 | 495.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 18 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 477.0 | 1.0 | 1.0 | 19.0 |
| 19 | 397.0 | 10.0 | 66.0 | 6.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 16.0 | 1.0 | 1.0 | 1.0 |
| 20 | 73.0 | 64.0 | 251.0 | 27.0 | 3.0 | 6.0 | 1.0 | 1.0 | 4.0 | 21.0 | 17.0 | 25.0 | 0.0 | 0.0 | 6.0 | 1.0 |
| 21 | 20.0 | 14.0 | 45.0 | 1.0 | 52.0 | 8.0 | 17.0 | 14.0 | 81.0 | 64.0 | 109.0 | 21.0 | 12.0 | 6.0 | 31.0 | 5.0 |
| 22 | 39.0 | 37.0 | 66.0 | 23.0 | 44.0 | 21.0 | 4.0 | 23.0 | 51.0 | 68.0 | 57.0 | 26.0 | 5.0 | 13.0 | 21.0 | 2.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.072 | -0.218 | 0.085 | -0.654 | 1.005 | 0.169 | -0.179 | -0.299 | -0.595 | 0.447 | 0.631 | -0.436 | -1.961 | -0.654 | 0.405 | -1.016 |
| 02 | -0.486 | -0.218 | 0.561 | 0.169 | 0.294 | -0.072 | -1.59 | 0.467 | -0.717 | 0.958 | 0.025 | 0.169 | -1.759 | -0.218 | -0.258 | -0.654 |
| 03 | -1.59 | -0.436 | 0.362 | -1.108 | -0.106 | 0.933 | -1.208 | 0.34 | -0.855 | 0.647 | 0.025 | -0.933 | -1.108 | 0.921 | 0.294 | -1.208 |
| 04 | -1.961 | -0.784 | -0.933 | -0.142 | -0.595 | -0.179 | -2.216 | 1.811 | -1.32 | 0.025 | -0.595 | 0.815 | -1.759 | -0.784 | 0.317 | -0.855 |
| 05 | -1.961 | -1.961 | -0.486 | -1.446 | -1.108 | -1.59 | -1.759 | 0.727 | -1.59 | -1.59 | -0.486 | 0.34 | -3.082 | -1.59 | 2.193 | -0.784 |
| 06 | -2.216 | -2.558 | -0.654 | -4.251 | -2.558 | -0.933 | -2.558 | -1.59 | -0.389 | 1.312 | 1.259 | 0.895 | -3.082 | -1.446 | 1.232 | -0.784 |
| 07 | -1.32 | -2.558 | -0.595 | -4.251 | 0.426 | -0.142 | -1.108 | 0.524 | 0.114 | 0.772 | 0.757 | 0.757 | -0.436 | -0.784 | 0.221 | -0.258 |
| 08 | -0.142 | -0.218 | 0.384 | -0.855 | 0.405 | 0.025 | -0.933 | -0.436 | 0.169 | 0.195 | 0.114 | -0.299 | -0.855 | -0.072 | 0.815 | -0.006 |
| 09 | 0.27 | -0.436 | 0.579 | -1.446 | 0.142 | 0.142 | -1.016 | 0.27 | 0.085 | 0.384 | 0.579 | -0.106 | -1.016 | -1.016 | 0.405 | -0.539 |
| 10 | -0.038 | -0.299 | 0.614 | -1.016 | -0.072 | -0.106 | -0.784 | 0.294 | 0.142 | 0.486 | 0.742 | -0.595 | -2.216 | -0.142 | 0.085 | -0.038 |
| 11 | -0.539 | -0.539 | 0.426 | -0.784 | -0.218 | 0.664 | -1.759 | 0.195 | -0.142 | 0.597 | 0.614 | -0.038 | -0.933 | -0.218 | 0.195 | -0.218 |
| 12 | -0.299 | -0.784 | 0.114 | -1.108 | 0.772 | 0.27 | -1.208 | 0.317 | 0.025 | 0.142 | 0.543 | -0.142 | -0.486 | -0.258 | 0.362 | -0.486 |
| 13 | -0.784 | 0.895 | 0.294 | -1.208 | -1.016 | 0.856 | -1.446 | -0.299 | -0.539 | 0.829 | 0.34 | -1.208 | -1.59 | 0.221 | 0.294 | -0.933 |
| 14 | 0.195 | -2.216 | -2.558 | -1.59 | 1.498 | -1.961 | -4.251 | 1.312 | 1.061 | -2.216 | -3.082 | 0.246 | -0.142 | -3.082 | -4.251 | -0.218 |
| 15 | -4.251 | -4.251 | 2.241 | -4.251 | -4.251 | -4.251 | -1.016 | -4.251 | -4.251 | -4.251 | -2.216 | -4.251 | -4.251 | -4.251 | 1.784 | -4.251 |
| 16 | -3.903 | -4.4 | -4.4 | -4.4 | -3.903 | -4.4 | -4.4 | -4.4 | 2.753 | -3.126 | -4.4 | -2.584 | -3.903 | -4.4 | -4.4 | -4.4 |
| 17 | -4.4 | -2.584 | -4.4 | 2.751 | -4.4 | -4.4 | -4.4 | -3.126 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.584 |
| 18 | -4.4 | -4.4 | -4.4 | -4.4 | -2.584 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.714 | -3.126 | -3.126 | -0.49 |
| 19 | 2.531 | -1.114 | 0.741 | -1.6 | -3.126 | -4.4 | -4.4 | -4.4 | -3.126 | -4.4 | -4.4 | -4.4 | -0.658 | -3.126 | -3.126 | -3.126 |
| 20 | 0.841 | 0.711 | 2.073 | -0.144 | -2.234 | -1.6 | -3.126 | -3.126 | -1.975 | -0.392 | -0.599 | -0.22 | -4.4 | -4.4 | -1.6 | -3.126 |
| 21 | -0.439 | -0.788 | 0.361 | -3.126 | 0.504 | -1.328 | -0.599 | -0.788 | 0.945 | 0.711 | 1.241 | -0.392 | -0.938 | -1.6 | -0.008 | -1.77 |
| 22 | 0.219 | 0.167 | 0.741 | -0.302 | 0.339 | -0.392 | -1.975 | -0.302 | 0.485 | 0.771 | 0.595 | -0.181 | -1.77 | -0.86 | -0.392 | -2.584 |