Model info
| Transcription factor | ZFP42 (GeneCards) | ||||||||
| Model | ZFP42_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 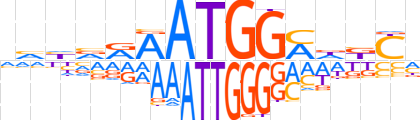 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bRvhKCCATTYbdbd | ||||||||
| Best auROC (human) | 0.938 | ||||||||
| Best auROC (mouse) | 0.84 | ||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 343 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | YY1-like factors {2.3.3.9} | ||||||||
| HGNC | HGNC:30949 | ||||||||
| EntrezGene | GeneID:132625 (SSTAR profile) | ||||||||
| UniProt ID | ZFP42_HUMAN | ||||||||
| UniProt AC | Q96MM3 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZFP42 expression | ||||||||
| ReMap ChIP-seq dataset list | ZFP42 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 6.0 | 1.0 | 33.0 | 1.0 | 2.0 | 14.0 | 26.0 | 5.0 | 12.0 | 5.0 | 83.0 | 6.0 | 5.0 | 0.0 | 97.0 | 3.0 |
| 02 | 4.0 | 13.0 | 8.0 | 0.0 | 13.0 | 5.0 | 2.0 | 0.0 | 117.0 | 86.0 | 26.0 | 10.0 | 5.0 | 8.0 | 0.0 | 2.0 |
| 03 | 7.0 | 6.0 | 1.0 | 125.0 | 69.0 | 28.0 | 10.0 | 5.0 | 20.0 | 14.0 | 1.0 | 1.0 | 3.0 | 6.0 | 2.0 | 1.0 |
| 04 | 0.0 | 0.0 | 97.0 | 2.0 | 0.0 | 0.0 | 35.0 | 19.0 | 0.0 | 0.0 | 14.0 | 0.0 | 1.0 | 0.0 | 2.0 | 129.0 |
| 05 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.0 | 134.0 | 1.0 | 1.0 | 2.0 | 148.0 | 0.0 | 0.0 |
| 06 | 0.0 | 14.0 | 0.0 | 0.0 | 0.0 | 283.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 296.0 | 2.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 296.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.0 | 28.0 | 21.0 | 241.0 |
| 10 | 2.0 | 1.0 | 2.0 | 4.0 | 2.0 | 2.0 | 0.0 | 24.0 | 4.0 | 9.0 | 1.0 | 7.0 | 8.0 | 111.0 | 6.0 | 116.0 |
| 11 | 0.0 | 8.0 | 1.0 | 7.0 | 4.0 | 55.0 | 57.0 | 7.0 | 0.0 | 1.0 | 2.0 | 6.0 | 2.0 | 16.0 | 15.0 | 118.0 |
| 12 | 1.0 | 1.0 | 1.0 | 3.0 | 55.0 | 7.0 | 6.0 | 12.0 | 73.0 | 1.0 | 1.0 | 0.0 | 26.0 | 13.0 | 67.0 | 32.0 |
| 13 | 8.0 | 12.0 | 30.0 | 105.0 | 3.0 | 6.0 | 3.0 | 10.0 | 14.0 | 8.0 | 29.0 | 24.0 | 6.0 | 5.0 | 22.0 | 14.0 |
| 14 | 13.0 | 6.0 | 7.0 | 5.0 | 8.0 | 11.0 | 4.0 | 8.0 | 23.0 | 12.0 | 28.0 | 21.0 | 9.0 | 9.0 | 29.0 | 106.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.097 | -2.642 | 0.561 | -2.642 | -2.09 | -0.283 | 0.325 | -1.268 | -0.433 | -1.268 | 1.476 | -1.097 | -1.268 | -3.979 | 1.632 | -1.736 |
| 02 | -1.475 | -0.355 | -0.824 | -3.979 | -0.355 | -1.268 | -2.09 | -3.979 | 1.818 | 1.512 | 0.325 | -0.609 | -1.268 | -0.824 | -3.979 | -2.09 |
| 03 | -0.951 | -1.097 | -2.642 | 1.884 | 1.293 | 0.398 | -0.609 | -1.268 | 0.067 | -0.283 | -2.642 | -2.642 | -1.736 | -1.097 | -2.09 | -2.642 |
| 04 | -3.979 | -3.979 | 1.632 | -2.09 | -3.979 | -3.979 | 0.619 | 0.016 | -3.979 | -3.979 | -0.283 | -3.979 | -2.642 | -3.979 | -2.09 | 1.916 |
| 05 | -3.979 | -2.642 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -0.433 | 1.954 | -2.642 | -2.642 | -2.09 | 2.053 | -3.979 | -3.979 |
| 06 | -3.979 | -0.283 | -3.979 | -3.979 | -3.979 | 2.7 | -3.979 | -3.979 | -3.979 | -2.642 | -3.979 | -3.979 | -3.979 | -2.642 | -3.979 | -3.979 |
| 07 | -3.979 | -3.979 | -3.979 | -3.979 | 2.745 | -2.09 | -3.979 | -2.642 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 |
| 08 | -3.979 | -3.979 | -3.979 | 2.745 | -3.979 | -3.979 | -3.979 | -2.09 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -2.642 |
| 09 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -3.979 | -0.711 | 0.398 | 0.115 | 2.54 |
| 10 | -2.09 | -2.642 | -2.09 | -1.475 | -2.09 | -2.09 | -3.979 | 0.246 | -1.475 | -0.711 | -2.642 | -0.951 | -0.824 | 1.766 | -1.097 | 1.81 |
| 11 | -3.979 | -0.824 | -2.642 | -0.951 | -1.475 | 1.067 | 1.103 | -0.951 | -3.979 | -2.642 | -2.09 | -1.097 | -2.09 | -0.152 | -0.215 | 1.827 |
| 12 | -2.642 | -2.642 | -2.642 | -1.736 | 1.067 | -0.951 | -1.097 | -0.433 | 1.349 | -2.642 | -2.642 | -3.979 | 0.325 | -0.355 | 1.263 | 0.53 |
| 13 | -0.824 | -0.433 | 0.466 | 1.711 | -1.736 | -1.097 | -1.736 | -0.609 | -0.283 | -0.824 | 0.433 | 0.246 | -1.097 | -1.268 | 0.16 | -0.283 |
| 14 | -0.355 | -1.097 | -0.951 | -1.268 | -0.824 | -0.517 | -1.475 | -0.824 | 0.204 | -0.433 | 0.398 | 0.115 | -0.711 | -0.711 | 0.433 | 1.72 |