Model info
| Transcription factor | AR (GeneCards) | ||||||||
| Model | ANDR_HUMAN.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 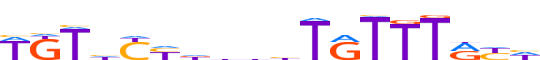 | ||||||||
| LOGO (reverse complement) | 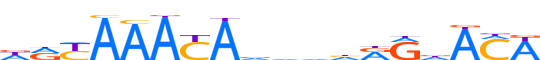 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | WKThYYddbhTRTTTRYh | ||||||||
| Best auROC (human) | 0.96 | ||||||||
| Best auROC (mouse) | 0.811 | ||||||||
| Peak sets in benchmark (human) | 461 | ||||||||
| Peak sets in benchmark (mouse) | 57 | ||||||||
| Aligned words | 499 | ||||||||
| TF family | Steroid hormone receptors (NR3) {2.1.1} | ||||||||
| TF subfamily | GR-like receptors (NR3C) {2.1.1.1} | ||||||||
| HGNC | HGNC:644 | ||||||||
| EntrezGene | GeneID:367 (SSTAR profile) | ||||||||
| UniProt ID | ANDR_HUMAN | ||||||||
| UniProt AC | P10275 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | AR expression | ||||||||
| ReMap ChIP-seq dataset list | AR datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 74.0 | 35.0 | 18.0 | 372.0 |
| 02 | 31.0 | 24.0 | 388.0 | 56.0 |
| 03 | 34.0 | 33.0 | 7.0 | 425.0 |
| 04 | 94.0 | 86.0 | 72.0 | 247.0 |
| 05 | 39.0 | 366.0 | 18.0 | 76.0 |
| 06 | 61.0 | 139.0 | 16.0 | 283.0 |
| 07 | 90.0 | 67.0 | 98.0 | 244.0 |
| 08 | 99.0 | 89.0 | 108.0 | 203.0 |
| 09 | 88.0 | 96.0 | 111.0 | 204.0 |
| 10 | 100.0 | 128.0 | 60.0 | 211.0 |
| 11 | 36.0 | 7.0 | 7.0 | 449.0 |
| 12 | 127.0 | 15.0 | 345.0 | 12.0 |
| 13 | 14.0 | 11.0 | 10.0 | 464.0 |
| 14 | 9.0 | 3.0 | 49.0 | 438.0 |
| 15 | 4.0 | 4.0 | 32.0 | 459.0 |
| 16 | 210.0 | 14.0 | 247.0 | 28.0 |
| 17 | 43.0 | 262.0 | 24.0 | 170.0 |
| 18 | 145.0 | 79.0 | 27.0 | 248.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.514 | -1.24 | -1.866 | 1.084 |
| 02 | -1.356 | -1.598 | 1.126 | -0.786 |
| 03 | -1.268 | -1.296 | -2.692 | 1.217 |
| 04 | -0.279 | -0.366 | -0.541 | 0.677 |
| 05 | -1.136 | 1.068 | -1.866 | -0.488 |
| 06 | -0.703 | 0.107 | -1.973 | 0.812 |
| 07 | -0.322 | -0.611 | -0.238 | 0.665 |
| 08 | -0.228 | -0.333 | -0.142 | 0.482 |
| 09 | -0.344 | -0.258 | -0.115 | 0.487 |
| 10 | -0.218 | 0.025 | -0.719 | 0.521 |
| 11 | -1.213 | -2.692 | -2.692 | 1.272 |
| 12 | 0.018 | -2.032 | 1.009 | -2.232 |
| 13 | -2.094 | -2.309 | -2.392 | 1.305 |
| 14 | -2.482 | -3.323 | -0.916 | 1.247 |
| 15 | -3.124 | -3.124 | -1.326 | 1.294 |
| 16 | 0.516 | -2.094 | 0.677 | -1.452 |
| 17 | -1.042 | 0.736 | -1.598 | 0.306 |
| 18 | 0.149 | -0.45 | -1.487 | 0.681 |