Model info
| Transcription factor | AR (GeneCards) | ||||||||
| Model | ANDR_HUMAN.H11MO.1.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 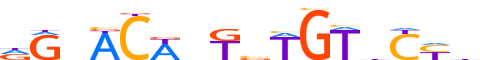 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 1 | ||||||||
| Consensus | dGnACWbTbWGTdCYb | ||||||||
| Best auROC (human) | 0.97 | ||||||||
| Best auROC (mouse) | 0.991 | ||||||||
| Peak sets in benchmark (human) | 461 | ||||||||
| Peak sets in benchmark (mouse) | 57 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Steroid hormone receptors (NR3) {2.1.1} | ||||||||
| TF subfamily | GR-like receptors (NR3C) {2.1.1.1} | ||||||||
| HGNC | HGNC:644 | ||||||||
| EntrezGene | GeneID:367 (SSTAR profile) | ||||||||
| UniProt ID | ANDR_HUMAN | ||||||||
| UniProt AC | P10275 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | AR expression | ||||||||
| ReMap ChIP-seq dataset list | AR datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 181.0 | 12.0 | 228.0 | 79.0 |
| 02 | 54.0 | 13.0 | 404.0 | 29.0 |
| 03 | 186.0 | 105.0 | 112.0 | 97.0 |
| 04 | 413.0 | 25.0 | 25.0 | 37.0 |
| 05 | 1.0 | 466.0 | 15.0 | 18.0 |
| 06 | 349.0 | 12.0 | 39.0 | 100.0 |
| 07 | 63.0 | 115.0 | 176.0 | 146.0 |
| 08 | 0.0 | 0.0 | 192.0 | 308.0 |
| 09 | 58.0 | 178.0 | 200.0 | 64.0 |
| 10 | 73.0 | 33.0 | 7.0 | 387.0 |
| 11 | 2.0 | 16.0 | 481.0 | 1.0 |
| 12 | 43.0 | 21.0 | 11.0 | 425.0 |
| 13 | 105.0 | 85.0 | 97.0 | 213.0 |
| 14 | 37.0 | 403.0 | 3.0 | 57.0 |
| 15 | 46.0 | 206.0 | 7.0 | 241.0 |
| 16 | 71.0 | 131.0 | 98.0 | 200.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.366 | -2.234 | 0.595 | -0.452 |
| 02 | -0.823 | -2.163 | 1.165 | -1.421 |
| 03 | 0.393 | -0.172 | -0.108 | -0.25 |
| 04 | 1.187 | -1.561 | -1.561 | -1.189 |
| 05 | -3.903 | 1.307 | -2.034 | -1.868 |
| 06 | 1.019 | -2.234 | -1.138 | -0.22 |
| 07 | -0.673 | -0.082 | 0.339 | 0.154 |
| 08 | -4.4 | -4.4 | 0.425 | 0.894 |
| 09 | -0.754 | 0.35 | 0.465 | -0.658 |
| 10 | -0.529 | -1.298 | -2.694 | 1.122 |
| 11 | -3.573 | -1.975 | 1.338 | -3.903 |
| 12 | -1.044 | -1.725 | -2.311 | 1.215 |
| 13 | -0.172 | -0.38 | -0.25 | 0.528 |
| 14 | -1.189 | 1.162 | -3.325 | -0.771 |
| 15 | -0.979 | 0.495 | -2.694 | 0.651 |
| 16 | -0.556 | 0.046 | -0.24 | 0.465 |