Model info
| Transcription factor | TFAP2C (GeneCards) | ||||||||
| Model | AP2C_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 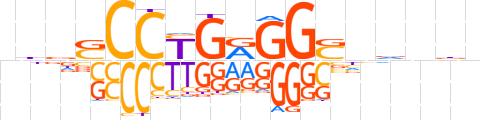 | ||||||||
| LOGO (reverse complement) | 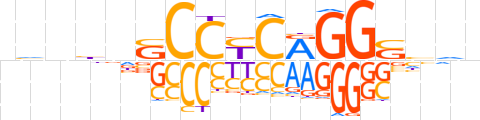 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nndSCCYGRGGShnndn | ||||||||
| Best auROC (human) | 0.964 | ||||||||
| Best auROC (mouse) | 0.959 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 6 | ||||||||
| Aligned words | 497 | ||||||||
| TF family | AP-2 {1.3.1} | ||||||||
| TF subfamily | AP-2gamma {1.3.1.0.3} | ||||||||
| HGNC | HGNC:11744 | ||||||||
| EntrezGene | GeneID:7022 (SSTAR profile) | ||||||||
| UniProt ID | AP2C_HUMAN | ||||||||
| UniProt AC | Q92754 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | TFAP2C expression | ||||||||
| ReMap ChIP-seq dataset list | TFAP2C datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 32.0 | 29.0 | 36.0 | 67.0 | 33.0 | 23.0 | 7.0 | 65.0 | 21.0 | 20.0 | 28.0 | 60.0 | 8.0 | 14.0 | 15.0 | 38.0 |
| 02 | 12.0 | 6.0 | 55.0 | 21.0 | 31.0 | 12.0 | 4.0 | 39.0 | 12.0 | 18.0 | 36.0 | 20.0 | 48.0 | 28.0 | 123.0 | 31.0 |
| 03 | 2.0 | 31.0 | 66.0 | 4.0 | 2.0 | 27.0 | 33.0 | 2.0 | 8.0 | 152.0 | 52.0 | 6.0 | 2.0 | 28.0 | 78.0 | 3.0 |
| 04 | 0.0 | 14.0 | 0.0 | 0.0 | 1.0 | 237.0 | 0.0 | 0.0 | 0.0 | 229.0 | 0.0 | 0.0 | 0.0 | 15.0 | 0.0 | 0.0 |
| 05 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 466.0 | 0.0 | 28.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 1.0 | 0.0 | 0.0 | 0.0 | 19.0 | 75.0 | 15.0 | 358.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 9.0 | 18.0 |
| 07 | 0.0 | 1.0 | 19.0 | 0.0 | 0.0 | 0.0 | 69.0 | 7.0 | 0.0 | 0.0 | 22.0 | 2.0 | 5.0 | 0.0 | 340.0 | 31.0 |
| 08 | 0.0 | 1.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 238.0 | 25.0 | 187.0 | 0.0 | 25.0 | 1.0 | 14.0 | 0.0 |
| 09 | 14.0 | 2.0 | 246.0 | 1.0 | 3.0 | 0.0 | 24.0 | 0.0 | 37.0 | 0.0 | 167.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 10 | 0.0 | 0.0 | 54.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 2.0 | 435.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| 11 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 17.0 | 302.0 | 159.0 | 14.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 12 | 3.0 | 4.0 | 4.0 | 7.0 | 86.0 | 69.0 | 43.0 | 105.0 | 22.0 | 96.0 | 12.0 | 31.0 | 3.0 | 1.0 | 4.0 | 6.0 |
| 13 | 53.0 | 18.0 | 29.0 | 14.0 | 95.0 | 23.0 | 13.0 | 39.0 | 19.0 | 17.0 | 14.0 | 13.0 | 59.0 | 17.0 | 64.0 | 9.0 |
| 14 | 38.0 | 44.0 | 68.0 | 76.0 | 18.0 | 24.0 | 2.0 | 31.0 | 25.0 | 21.0 | 35.0 | 39.0 | 7.0 | 18.0 | 27.0 | 23.0 |
| 15 | 24.0 | 7.0 | 37.0 | 20.0 | 33.0 | 14.0 | 20.0 | 40.0 | 36.0 | 14.0 | 45.0 | 37.0 | 15.0 | 19.0 | 72.0 | 63.0 |
| 16 | 18.0 | 19.0 | 49.0 | 22.0 | 14.0 | 20.0 | 4.0 | 16.0 | 21.0 | 51.0 | 72.0 | 30.0 | 17.0 | 42.0 | 48.0 | 53.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.031 | -0.066 | 0.148 | 0.764 | 0.062 | -0.294 | -1.447 | 0.734 | -0.384 | -0.431 | -0.1 | 0.654 | -1.32 | -0.78 | -0.713 | 0.201 |
| 02 | -0.93 | -1.592 | 0.568 | -0.384 | 0.0 | -0.93 | -1.968 | 0.227 | -0.93 | -0.535 | 0.148 | -0.431 | 0.433 | -0.1 | 1.369 | 0.0 |
| 03 | -2.576 | 0.0 | 0.749 | -1.968 | -2.576 | -0.136 | 0.062 | -2.576 | -1.32 | 1.58 | 0.512 | -1.592 | -2.576 | -0.1 | 0.915 | -2.226 |
| 04 | -4.393 | -0.78 | -4.393 | -4.393 | -3.119 | 2.023 | -4.393 | -4.393 | -4.393 | 1.989 | -4.393 | -4.393 | -4.393 | -0.713 | -4.393 | -4.393 |
| 05 | -4.393 | -3.119 | -4.393 | -4.393 | -3.119 | 2.699 | -4.393 | -0.1 | -4.393 | -4.393 | -4.393 | -4.393 | -4.393 | -4.393 | -4.393 | -4.393 |
| 06 | -3.119 | -4.393 | -4.393 | -4.393 | -0.482 | 0.876 | -0.713 | 2.435 | -4.393 | -4.393 | -4.393 | -4.393 | -4.393 | -3.119 | -1.207 | -0.535 |
| 07 | -4.393 | -3.119 | -0.482 | -4.393 | -4.393 | -4.393 | 0.793 | -1.447 | -4.393 | -4.393 | -0.338 | -2.576 | -1.762 | -4.393 | 2.384 | 0.0 |
| 08 | -4.393 | -3.119 | -1.968 | -4.393 | -4.393 | -4.393 | -4.393 | -3.119 | 2.027 | -0.212 | 1.787 | -4.393 | -0.212 | -3.119 | -0.78 | -4.393 |
| 09 | -0.78 | -2.576 | 2.06 | -3.119 | -2.226 | -4.393 | -0.252 | -4.393 | 0.175 | -4.393 | 1.674 | -3.119 | -4.393 | -4.393 | -3.119 | -4.393 |
| 10 | -4.393 | -4.393 | 0.55 | -4.393 | -4.393 | -4.393 | -2.576 | -4.393 | -4.393 | -2.576 | 2.63 | -3.119 | -3.119 | -4.393 | -3.119 | -4.393 |
| 11 | -4.393 | -4.393 | -3.119 | -4.393 | -3.119 | -3.119 | -4.393 | -4.393 | -0.591 | 2.265 | 1.625 | -0.78 | -4.393 | -4.393 | -3.119 | -4.393 |
| 12 | -2.226 | -1.968 | -1.968 | -1.447 | 1.012 | 0.793 | 0.324 | 1.211 | -0.338 | 1.122 | -0.93 | 0.0 | -2.226 | -3.119 | -1.968 | -1.592 |
| 13 | 0.531 | -0.535 | -0.066 | -0.78 | 1.112 | -0.294 | -0.852 | 0.227 | -0.482 | -0.591 | -0.78 | -0.852 | 0.638 | -0.591 | 0.719 | -1.207 |
| 14 | 0.201 | 0.347 | 0.779 | 0.889 | -0.535 | -0.252 | -2.576 | 0.0 | -0.212 | -0.384 | 0.12 | 0.227 | -1.447 | -0.535 | -0.136 | -0.294 |
| 15 | -0.252 | -1.447 | 0.175 | -0.431 | 0.062 | -0.78 | -0.431 | 0.252 | 0.148 | -0.78 | 0.369 | 0.175 | -0.713 | -0.482 | 0.836 | 0.703 |
| 16 | -0.535 | -0.482 | 0.453 | -0.338 | -0.78 | -0.431 | -1.968 | -0.65 | -0.384 | 0.493 | 0.836 | -0.032 | -0.591 | 0.3 | 0.433 | 0.531 |