Model info
| Transcription factor | ASCL1 (GeneCards) | ||||||||
| Model | ASCL1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 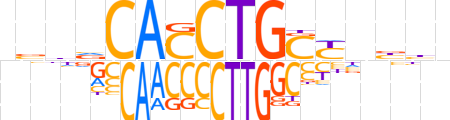 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvhvCACCTGSYnbhn | ||||||||
| Best auROC (human) | 0.99 | ||||||||
| Best auROC (mouse) | 0.983 | ||||||||
| Peak sets in benchmark (human) | 30 | ||||||||
| Peak sets in benchmark (mouse) | 25 | ||||||||
| Aligned words | 534 | ||||||||
| TF family | MyoD / ASC-related factors {1.2.2} | ||||||||
| TF subfamily | Achaete-Scute-like factors {1.2.2.2} | ||||||||
| HGNC | HGNC:738 | ||||||||
| EntrezGene | GeneID:429 (SSTAR profile) | ||||||||
| UniProt ID | ASCL1_HUMAN | ||||||||
| UniProt AC | P50553 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ASCL1 expression | ||||||||
| ReMap ChIP-seq dataset list | ASCL1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 25.0 | 21.0 | 25.0 | 7.0 | 16.0 | 88.0 | 14.0 | 26.0 | 19.0 | 66.0 | 35.0 | 12.0 | 12.0 | 47.0 | 55.0 | 16.0 |
| 02 | 23.0 | 28.0 | 7.0 | 14.0 | 61.0 | 40.0 | 13.0 | 108.0 | 28.0 | 46.0 | 20.0 | 35.0 | 12.0 | 11.0 | 18.0 | 20.0 |
| 03 | 33.0 | 13.0 | 68.0 | 10.0 | 59.0 | 22.0 | 20.0 | 24.0 | 8.0 | 35.0 | 14.0 | 1.0 | 11.0 | 17.0 | 137.0 | 12.0 |
| 04 | 0.0 | 111.0 | 0.0 | 0.0 | 0.0 | 87.0 | 0.0 | 0.0 | 0.0 | 238.0 | 0.0 | 1.0 | 0.0 | 47.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 483.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 365.0 | 119.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 365.0 | 0.0 | 0.0 | 2.0 | 117.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 482.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 483.0 | 1.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 372.0 | 55.0 | 56.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 21.0 | 189.0 | 8.0 | 154.0 | 4.0 | 47.0 | 0.0 | 5.0 | 3.0 | 37.0 | 1.0 | 15.0 |
| 12 | 9.0 | 9.0 | 4.0 | 6.0 | 86.0 | 69.0 | 18.0 | 100.0 | 0.0 | 1.0 | 3.0 | 5.0 | 13.0 | 69.0 | 65.0 | 27.0 |
| 13 | 7.0 | 54.0 | 26.0 | 21.0 | 22.0 | 65.0 | 14.0 | 47.0 | 2.0 | 57.0 | 18.0 | 13.0 | 8.0 | 65.0 | 36.0 | 29.0 |
| 14 | 5.0 | 19.0 | 7.0 | 8.0 | 50.0 | 83.0 | 12.0 | 96.0 | 5.0 | 40.0 | 21.0 | 28.0 | 4.0 | 49.0 | 23.0 | 34.0 |
| 15 | 6.0 | 14.0 | 31.0 | 13.0 | 44.0 | 75.0 | 12.0 | 60.0 | 9.0 | 26.0 | 14.0 | 14.0 | 12.0 | 54.0 | 62.0 | 38.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.188 | -0.359 | -0.188 | -1.423 | -0.626 | 1.06 | -0.756 | -0.149 | -0.458 | 0.773 | 0.144 | -0.906 | -0.906 | 0.436 | 0.592 | -0.626 |
| 02 | -0.27 | -0.076 | -1.423 | -0.756 | 0.695 | 0.276 | -0.828 | 1.264 | -0.076 | 0.415 | -0.407 | 0.144 | -0.906 | -0.99 | -0.511 | -0.407 |
| 03 | 0.086 | -0.828 | 0.803 | -1.082 | 0.662 | -0.314 | -0.407 | -0.228 | -1.296 | 0.144 | -0.756 | -3.095 | -0.99 | -0.567 | 1.501 | -0.906 |
| 04 | -4.373 | 1.291 | -4.373 | -4.373 | -4.373 | 1.048 | -4.373 | -4.373 | -4.373 | 2.052 | -4.373 | -3.095 | -4.373 | 0.436 | -4.373 | -4.373 |
| 05 | -4.373 | -4.373 | -4.373 | -4.373 | 2.759 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -3.095 | -4.373 | -4.373 | -4.373 |
| 06 | -4.373 | 2.479 | 1.36 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 |
| 07 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | 2.479 | -4.373 | -4.373 | -2.552 | 1.343 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 |
| 08 | -4.373 | -4.373 | -4.373 | -2.552 | -4.373 | -4.373 | -4.373 | 2.757 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 |
| 09 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | 2.759 | -3.095 |
| 10 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | -4.373 | 2.498 | 0.592 | 0.61 | -4.373 | -4.373 | -3.095 | -4.373 |
| 11 | -4.373 | -4.373 | -4.373 | -4.373 | -0.359 | 1.822 | -1.296 | 1.617 | -1.944 | 0.436 | -4.373 | -1.738 | -2.202 | 0.199 | -3.095 | -0.689 |
| 12 | -1.183 | -1.183 | -1.944 | -1.568 | 1.037 | 0.818 | -0.511 | 1.187 | -4.373 | -3.095 | -2.202 | -1.738 | -0.828 | 0.818 | 0.758 | -0.112 |
| 13 | -1.423 | 0.574 | -0.149 | -0.359 | -0.314 | 0.758 | -0.756 | 0.436 | -2.552 | 0.628 | -0.511 | -0.828 | -1.296 | 0.758 | 0.172 | -0.042 |
| 14 | -1.738 | -0.458 | -1.423 | -1.296 | 0.498 | 1.001 | -0.906 | 1.146 | -1.738 | 0.276 | -0.359 | -0.076 | -1.944 | 0.477 | -0.27 | 0.115 |
| 15 | -1.568 | -0.756 | 0.024 | -0.828 | 0.371 | 0.9 | -0.906 | 0.679 | -1.183 | -0.149 | -0.756 | -0.756 | -0.906 | 0.574 | 0.711 | 0.226 |