Model info
| Transcription factor | ATF1 (GeneCards) | ||||||||
| Model | ATF1_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 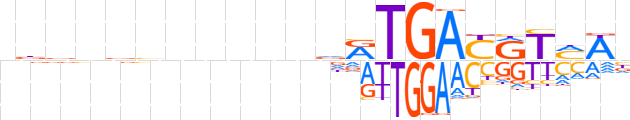 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 22 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndbnvvnnnvndRTGAYRTMWn | ||||||||
| Best auROC (human) | 0.867 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 6 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 505 | ||||||||
| TF family | CREB-related factors {1.1.7} | ||||||||
| TF subfamily | CREB-like factors {1.1.7.1} | ||||||||
| HGNC | HGNC:783 | ||||||||
| EntrezGene | GeneID:466 (SSTAR profile) | ||||||||
| UniProt ID | ATF1_HUMAN | ||||||||
| UniProt AC | P18846 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ATF1 expression | ||||||||
| ReMap ChIP-seq dataset list | ATF1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 23.085 | 8.345 | 54.017 | 31.881 | 23.104 | 23.416 | 75.21 | 31.76 | 27.587 | 16.421 | 55.877 | 39.858 | 14.581 | 19.553 | 29.538 | 25.766 |
| 02 | 26.973 | 13.743 | 30.99 | 16.651 | 22.931 | 18.012 | 13.418 | 13.375 | 26.092 | 39.059 | 109.133 | 40.357 | 4.266 | 26.107 | 54.85 | 44.043 |
| 03 | 24.13 | 20.849 | 23.191 | 12.092 | 24.132 | 12.969 | 19.187 | 40.633 | 31.602 | 114.014 | 31.336 | 31.44 | 7.676 | 50.082 | 29.503 | 27.165 |
| 04 | 30.397 | 15.469 | 27.953 | 13.721 | 35.387 | 35.449 | 110.147 | 16.932 | 28.163 | 24.829 | 34.988 | 15.237 | 13.329 | 33.168 | 36.039 | 28.794 |
| 05 | 14.742 | 37.725 | 39.049 | 15.759 | 40.522 | 26.992 | 15.111 | 26.29 | 26.573 | 126.798 | 43.787 | 11.969 | 17.112 | 13.833 | 24.956 | 18.781 |
| 06 | 18.205 | 30.944 | 27.757 | 22.043 | 52.682 | 56.553 | 60.275 | 35.838 | 36.228 | 30.147 | 31.699 | 24.829 | 9.649 | 19.729 | 16.909 | 26.513 |
| 07 | 33.096 | 35.126 | 36.194 | 12.348 | 36.219 | 41.199 | 21.368 | 38.588 | 25.705 | 54.11 | 28.987 | 27.837 | 13.343 | 33.939 | 42.147 | 19.794 |
| 08 | 27.978 | 18.737 | 54.855 | 6.792 | 38.823 | 57.468 | 37.581 | 30.501 | 25.823 | 31.696 | 47.316 | 23.861 | 9.069 | 25.446 | 39.258 | 24.794 |
| 09 | 24.383 | 20.548 | 48.349 | 8.414 | 34.508 | 48.488 | 26.638 | 23.714 | 32.28 | 61.295 | 66.582 | 18.853 | 12.071 | 28.689 | 27.442 | 17.746 |
| 10 | 31.792 | 18.213 | 22.051 | 31.186 | 27.995 | 46.878 | 20.061 | 64.086 | 58.785 | 44.739 | 22.18 | 43.306 | 14.288 | 18.72 | 18.735 | 16.985 |
| 11 | 58.47 | 12.716 | 48.575 | 13.099 | 43.246 | 14.924 | 24.641 | 45.74 | 33.427 | 11.462 | 27.562 | 10.576 | 20.562 | 8.834 | 90.92 | 35.247 |
| 12 | 86.483 | 6.245 | 62.976 | 0.0 | 32.709 | 0.0 | 15.227 | 0.0 | 121.429 | 16.255 | 52.029 | 1.986 | 50.028 | 12.745 | 41.122 | 0.766 |
| 13 | 5.352 | 0.0 | 0.746 | 284.551 | 0.818 | 1.192 | 0.0 | 33.234 | 1.218 | 1.221 | 0.0 | 168.915 | 0.0 | 0.0 | 0.0 | 2.752 |
| 14 | 0.0 | 0.0 | 6.278 | 1.111 | 0.0 | 0.0 | 2.413 | 0.0 | 0.0 | 0.0 | 0.746 | 0.0 | 1.76 | 0.0 | 486.438 | 1.253 |
| 15 | 1.76 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 472.674 | 1.171 | 15.429 | 6.601 | 1.253 | 0.0 | 1.111 | 0.0 |
| 16 | 16.89 | 348.331 | 21.531 | 88.936 | 0.0 | 1.171 | 0.0 | 0.0 | 1.111 | 13.703 | 0.0 | 1.726 | 0.719 | 4.34 | 0.0 | 1.542 |
| 17 | 5.235 | 2.928 | 10.558 | 0.0 | 47.192 | 23.206 | 263.943 | 33.204 | 2.883 | 1.496 | 15.224 | 1.927 | 1.624 | 4.06 | 85.643 | 0.876 |
| 18 | 1.828 | 2.065 | 4.023 | 49.018 | 0.0 | 1.587 | 0.0 | 30.103 | 6.852 | 61.473 | 3.051 | 303.993 | 5.317 | 20.029 | 0.79 | 9.871 |
| 19 | 4.973 | 7.961 | 0.0 | 1.062 | 70.116 | 5.81 | 3.732 | 5.497 | 0.772 | 6.345 | 0.746 | 0.0 | 97.538 | 244.574 | 23.251 | 27.623 |
| 20 | 115.424 | 8.961 | 28.36 | 20.654 | 241.134 | 0.0 | 8.629 | 14.928 | 16.51 | 1.519 | 3.645 | 6.055 | 13.162 | 4.179 | 1.426 | 15.416 |
| 21 | 65.866 | 114.634 | 95.806 | 109.924 | 3.998 | 1.707 | 4.675 | 4.278 | 15.717 | 13.078 | 3.461 | 9.804 | 15.948 | 19.929 | 9.08 | 12.095 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.298 | -1.287 | 0.542 | 0.02 | -0.298 | -0.284 | 0.871 | 0.016 | -0.123 | -0.632 | 0.576 | 0.241 | -0.748 | -0.462 | -0.056 | -0.19 |
| 02 | -0.145 | -0.806 | -0.008 | -0.619 | -0.305 | -0.542 | -0.829 | -0.832 | -0.178 | 0.221 | 1.242 | 0.253 | -1.917 | -0.177 | 0.557 | 0.34 |
| 03 | -0.255 | -0.399 | -0.294 | -0.93 | -0.255 | -0.862 | -0.48 | 0.26 | 0.011 | 1.285 | 0.003 | 0.006 | -1.367 | 0.467 | -0.057 | -0.138 |
| 04 | -0.027 | -0.691 | -0.11 | -0.808 | 0.123 | 0.125 | 1.251 | -0.602 | -0.103 | -0.227 | 0.112 | -0.706 | -0.836 | 0.059 | 0.141 | -0.081 |
| 05 | -0.738 | 0.186 | 0.22 | -0.673 | 0.257 | -0.145 | -0.714 | -0.171 | -0.16 | 1.391 | 0.334 | -0.94 | -0.592 | -0.8 | -0.222 | -0.501 |
| 06 | -0.532 | -0.01 | -0.117 | -0.344 | 0.517 | 0.588 | 0.651 | 0.135 | 0.146 | -0.035 | 0.014 | -0.227 | -1.148 | -0.453 | -0.604 | -0.162 |
| 07 | 0.057 | 0.116 | 0.145 | -0.91 | 0.146 | 0.273 | -0.374 | 0.209 | -0.193 | 0.544 | -0.074 | -0.114 | -0.835 | 0.082 | 0.296 | -0.45 |
| 08 | -0.109 | -0.503 | 0.557 | -1.483 | 0.215 | 0.604 | 0.182 | -0.024 | -0.188 | 0.014 | 0.411 | -0.266 | -1.208 | -0.203 | 0.226 | -0.228 |
| 09 | -0.245 | -0.413 | 0.432 | -1.279 | 0.098 | 0.435 | -0.158 | -0.272 | 0.032 | 0.668 | 0.75 | -0.497 | -0.932 | -0.084 | -0.128 | -0.557 |
| 10 | 0.017 | -0.531 | -0.344 | -0.002 | -0.109 | 0.401 | -0.436 | 0.712 | 0.626 | 0.355 | -0.338 | 0.323 | -0.768 | -0.504 | -0.503 | -0.599 |
| 11 | 0.621 | -0.881 | 0.437 | -0.853 | 0.321 | -0.726 | -0.234 | 0.377 | 0.067 | -0.982 | -0.124 | -1.06 | -0.412 | -1.233 | 1.06 | 0.119 |
| 12 | 1.01 | -1.562 | 0.695 | -4.4 | 0.045 | -4.4 | -0.706 | -4.4 | 1.348 | -0.642 | 0.505 | -2.59 | 0.466 | -0.879 | 0.272 | -3.311 |
| 13 | -1.707 | -4.4 | -3.328 | 2.198 | -3.266 | -2.997 | -4.4 | 0.061 | -2.98 | -2.978 | -4.4 | 1.677 | -4.4 | -4.4 | -4.4 | -2.31 |
| 14 | -4.4 | -4.4 | -1.557 | -3.049 | -4.4 | -4.4 | -2.424 | -4.4 | -4.4 | -4.4 | -3.328 | -4.4 | -2.689 | -4.4 | 2.734 | -2.959 |
| 15 | -2.689 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.705 | -3.01 | -0.693 | -1.51 | -2.959 | -4.4 | -3.049 | -4.4 |
| 16 | -0.605 | 2.4 | -0.367 | 1.038 | -4.4 | -3.01 | -4.4 | -4.4 | -3.049 | -0.809 | -4.4 | -2.706 | -3.352 | -1.901 | -4.4 | -2.797 |
| 17 | -1.727 | -2.256 | -1.061 | -4.4 | 0.408 | -0.293 | 2.123 | 0.06 | -2.269 | -2.821 | -0.706 | -2.615 | -2.755 | -1.962 | 1.0 | -3.22 |
| 18 | -2.659 | -2.557 | -1.97 | 0.446 | -4.4 | -2.773 | -4.4 | -0.037 | -1.475 | 0.671 | -2.219 | 2.264 | -1.713 | -0.438 | -3.29 | -1.126 |
| 19 | -1.775 | -1.332 | -4.4 | -3.083 | 0.801 | -1.63 | -2.038 | -1.682 | -3.305 | -1.547 | -3.328 | -4.4 | 1.13 | 2.047 | -0.291 | -0.122 |
| 20 | 1.298 | -1.219 | -0.096 | -0.408 | 2.033 | -4.4 | -1.255 | -0.725 | -0.627 | -2.809 | -2.06 | -1.591 | -0.848 | -1.936 | -2.859 | -0.694 |
| 21 | 0.739 | 1.291 | 1.112 | 1.249 | -1.976 | -2.714 | -1.832 | -1.914 | -0.675 | -0.854 | -2.107 | -1.133 | -0.661 | -0.443 | -1.206 | -0.93 |