Model info
| Transcription factor | Barx1 | ||||||||
| Model | BARX1_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 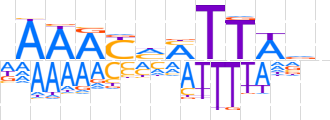 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbWAAKKRTTTh | ||||||||
| Best auROC (human) | 0.566 | ||||||||
| Best auROC (mouse) | 0.832 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 462 | ||||||||
| TF family | NK-related factors {3.1.2} | ||||||||
| TF subfamily | BARX {3.1.2.2} | ||||||||
| MGI | MGI:103124 | ||||||||
| EntrezGene | GeneID:12022 (SSTAR profile) | ||||||||
| UniProt ID | BARX1_MOUSE | ||||||||
| UniProt AC | Q9ER42 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Barx1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 9.0 | 44.0 | 28.0 | 30.0 | 10.0 | 36.0 | 4.0 | 44.0 | 16.0 | 44.0 | 20.0 | 26.0 | 9.0 | 42.0 | 44.0 | 55.0 |
| 02 | 13.0 | 0.0 | 0.0 | 31.0 | 50.0 | 3.0 | 0.0 | 113.0 | 24.0 | 4.0 | 8.0 | 60.0 | 30.0 | 2.0 | 3.0 | 120.0 |
| 03 | 102.0 | 11.0 | 2.0 | 2.0 | 6.0 | 2.0 | 0.0 | 1.0 | 11.0 | 0.0 | 0.0 | 0.0 | 299.0 | 14.0 | 3.0 | 8.0 |
| 04 | 414.0 | 1.0 | 2.0 | 1.0 | 26.0 | 0.0 | 1.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 7.0 | 2.0 | 1.0 | 1.0 |
| 05 | 28.0 | 24.0 | 87.0 | 313.0 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 | 0.0 | 1.0 | 3.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 06 | 5.0 | 4.0 | 2.0 | 17.0 | 6.0 | 2.0 | 6.0 | 12.0 | 9.0 | 1.0 | 19.0 | 60.0 | 33.0 | 10.0 | 173.0 | 102.0 |
| 07 | 2.0 | 3.0 | 45.0 | 3.0 | 0.0 | 2.0 | 14.0 | 1.0 | 27.0 | 22.0 | 130.0 | 21.0 | 21.0 | 8.0 | 161.0 | 1.0 |
| 08 | 0.0 | 0.0 | 1.0 | 49.0 | 0.0 | 2.0 | 1.0 | 32.0 | 5.0 | 63.0 | 13.0 | 269.0 | 0.0 | 3.0 | 3.0 | 20.0 |
| 09 | 3.0 | 0.0 | 0.0 | 2.0 | 10.0 | 7.0 | 0.0 | 51.0 | 1.0 | 0.0 | 1.0 | 16.0 | 26.0 | 8.0 | 15.0 | 321.0 |
| 10 | 2.0 | 0.0 | 0.0 | 38.0 | 0.0 | 2.0 | 0.0 | 13.0 | 1.0 | 0.0 | 4.0 | 11.0 | 19.0 | 5.0 | 12.0 | 354.0 |
| 11 | 7.0 | 5.0 | 4.0 | 6.0 | 1.0 | 1.0 | 0.0 | 5.0 | 5.0 | 1.0 | 7.0 | 3.0 | 106.0 | 97.0 | 37.0 | 176.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.135 | 0.419 | -0.028 | 0.04 | -1.034 | 0.22 | -1.896 | 0.419 | -0.578 | 0.419 | -0.359 | -0.101 | -1.135 | 0.373 | 0.419 | 0.64 |
| 02 | -0.78 | -4.333 | -4.333 | 0.072 | 0.546 | -2.155 | -4.333 | 1.357 | -0.18 | -1.896 | -1.248 | 0.727 | 0.04 | -2.506 | -2.155 | 1.417 |
| 03 | 1.255 | -0.942 | -2.506 | -2.506 | -1.52 | -2.506 | -4.333 | -3.05 | -0.942 | -4.333 | -4.333 | -4.333 | 2.328 | -0.708 | -2.155 | -1.248 |
| 04 | 2.653 | -3.05 | -2.506 | -3.05 | -0.101 | -4.333 | -3.05 | -4.333 | -1.691 | -4.333 | -4.333 | -4.333 | -1.375 | -2.506 | -3.05 | -3.05 |
| 05 | -0.028 | -0.18 | 1.096 | 2.373 | -4.333 | -3.05 | -4.333 | -2.506 | -4.333 | -4.333 | -3.05 | -2.155 | -4.333 | -3.05 | -3.05 | -4.333 |
| 06 | -1.691 | -1.896 | -2.506 | -0.519 | -1.52 | -2.506 | -1.52 | -0.858 | -1.135 | -3.05 | -0.41 | 0.727 | 0.134 | -1.034 | 1.781 | 1.255 |
| 07 | -2.506 | -2.155 | 0.441 | -2.155 | -4.333 | -2.506 | -0.708 | -3.05 | -0.064 | -0.266 | 1.496 | -0.311 | -0.311 | -1.248 | 1.71 | -3.05 |
| 08 | -4.333 | -4.333 | -3.05 | 0.526 | -4.333 | -2.506 | -3.05 | 0.104 | -1.691 | 0.775 | -0.78 | 2.222 | -4.333 | -2.155 | -2.155 | -0.359 |
| 09 | -2.155 | -4.333 | -4.333 | -2.506 | -1.034 | -1.375 | -4.333 | 0.565 | -3.05 | -4.333 | -3.05 | -0.578 | -0.101 | -1.248 | -0.641 | 2.399 |
| 10 | -2.506 | -4.333 | -4.333 | 0.274 | -4.333 | -2.506 | -4.333 | -0.78 | -3.05 | -4.333 | -1.896 | -0.942 | -0.41 | -1.691 | -0.858 | 2.496 |
| 11 | -1.375 | -1.691 | -1.896 | -1.52 | -3.05 | -3.05 | -4.333 | -1.691 | -1.691 | -3.05 | -1.375 | -2.155 | 1.293 | 1.205 | 0.247 | 1.799 |