Model info
| Transcription factor | Barx1 | ||||||||
| Model | BARX1_MOUSE.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 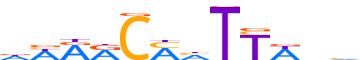 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bbWAAdKGYYWh | ||||||||
| Best auROC (human) | 0.529 | ||||||||
| Best auROC (mouse) | 0.819 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 150 | ||||||||
| TF family | NK-related factors {3.1.2} | ||||||||
| TF subfamily | BARX {3.1.2.2} | ||||||||
| MGI | MGI:103124 | ||||||||
| EntrezGene | GeneID:12022 (SSTAR profile) | ||||||||
| UniProt ID | BARX1_MOUSE | ||||||||
| UniProt AC | Q9ER42 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Barx1 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 20.0 | 28.0 | 50.0 | 52.0 |
| 02 | 21.0 | 49.0 | 28.0 | 52.0 |
| 03 | 30.0 | 18.0 | 7.0 | 95.0 |
| 04 | 119.0 | 12.0 | 18.0 | 1.0 |
| 05 | 146.0 | 3.0 | 0.0 | 1.0 |
| 06 | 19.0 | 15.0 | 39.0 | 77.0 |
| 07 | 15.0 | 1.0 | 45.0 | 89.0 |
| 08 | 2.0 | 8.0 | 139.0 | 1.0 |
| 09 | 12.0 | 43.0 | 6.0 | 89.0 |
| 10 | 22.0 | 24.0 | 2.0 | 102.0 |
| 11 | 22.0 | 16.0 | 18.0 | 94.0 |
| 12 | 28.0 | 26.0 | 18.0 | 78.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.601 | -0.281 | 0.28 | 0.318 |
| 02 | -0.555 | 0.26 | -0.281 | 0.318 |
| 03 | -0.215 | -0.7 | -1.547 | 0.91 |
| 04 | 1.132 | -1.073 | -0.7 | -2.845 |
| 05 | 1.335 | -2.21 | -3.432 | -2.845 |
| 06 | -0.649 | -0.869 | 0.038 | 0.703 |
| 07 | -0.869 | -2.845 | 0.177 | 0.845 |
| 08 | -2.478 | -1.432 | 1.286 | -2.845 |
| 09 | -1.073 | 0.133 | -1.676 | 0.845 |
| 10 | -0.511 | -0.428 | -2.478 | 0.98 |
| 11 | -0.511 | -0.809 | -0.7 | 0.899 |
| 12 | -0.281 | -0.352 | -0.7 | 0.715 |