Model info
| Transcription factor | Arntl | ||||||||
| Model | BMAL1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 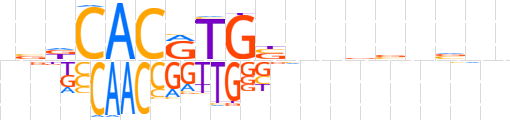 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nnbbvbnnSCAYGTGhbn | ||||||||
| Best auROC (human) | 0.859 | ||||||||
| Best auROC (mouse) | 0.883 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 15 | ||||||||
| Aligned words | 420 | ||||||||
| TF family | PAS domain factors {1.2.5} | ||||||||
| TF subfamily | Arnt-like factors {1.2.5.2} | ||||||||
| MGI | MGI:1096381 | ||||||||
| EntrezGene | GeneID:11865 (SSTAR profile) | ||||||||
| UniProt ID | BMAL1_MOUSE | ||||||||
| UniProt AC | Q9WTL8 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Arntl expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 8.0 | 13.0 | 18.0 | 31.0 | 33.0 | 24.0 | 38.0 | 35.0 | 19.0 | 34.0 | 12.0 | 59.0 | 8.0 | 11.0 | 51.0 | 16.0 |
| 02 | 9.0 | 15.0 | 33.0 | 11.0 | 16.0 | 22.0 | 17.0 | 27.0 | 9.0 | 41.0 | 48.0 | 21.0 | 8.0 | 29.0 | 91.0 | 13.0 |
| 03 | 4.0 | 10.0 | 21.0 | 7.0 | 22.0 | 47.0 | 14.0 | 24.0 | 19.0 | 61.0 | 58.0 | 51.0 | 5.0 | 24.0 | 33.0 | 10.0 |
| 04 | 2.0 | 19.0 | 22.0 | 7.0 | 32.0 | 58.0 | 32.0 | 20.0 | 18.0 | 44.0 | 48.0 | 16.0 | 10.0 | 23.0 | 52.0 | 7.0 |
| 05 | 11.0 | 26.0 | 16.0 | 9.0 | 23.0 | 58.0 | 20.0 | 43.0 | 23.0 | 64.0 | 26.0 | 41.0 | 3.0 | 15.0 | 13.0 | 19.0 |
| 06 | 10.0 | 6.0 | 42.0 | 2.0 | 52.0 | 30.0 | 35.0 | 46.0 | 10.0 | 23.0 | 26.0 | 16.0 | 15.0 | 15.0 | 52.0 | 30.0 |
| 07 | 21.0 | 16.0 | 28.0 | 22.0 | 25.0 | 13.0 | 14.0 | 22.0 | 16.0 | 45.0 | 46.0 | 48.0 | 19.0 | 11.0 | 45.0 | 19.0 |
| 08 | 20.0 | 24.0 | 36.0 | 1.0 | 30.0 | 39.0 | 15.0 | 1.0 | 32.0 | 73.0 | 27.0 | 1.0 | 14.0 | 60.0 | 37.0 | 0.0 |
| 09 | 6.0 | 89.0 | 1.0 | 0.0 | 9.0 | 186.0 | 0.0 | 1.0 | 4.0 | 107.0 | 2.0 | 2.0 | 0.0 | 1.0 | 0.0 | 2.0 |
| 10 | 19.0 | 0.0 | 0.0 | 0.0 | 350.0 | 3.0 | 18.0 | 12.0 | 2.0 | 1.0 | 0.0 | 0.0 | 4.0 | 0.0 | 1.0 | 0.0 |
| 11 | 7.0 | 261.0 | 23.0 | 84.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 17.0 | 0.0 | 2.0 | 0.0 | 9.0 | 0.0 | 3.0 |
| 12 | 0.0 | 1.0 | 6.0 | 0.0 | 1.0 | 2.0 | 285.0 | 3.0 | 1.0 | 0.0 | 22.0 | 0.0 | 1.0 | 1.0 | 84.0 | 3.0 |
| 13 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 4.0 | 3.0 | 0.0 | 3.0 | 391.0 | 0.0 | 0.0 | 0.0 | 6.0 |
| 14 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 391.0 | 13.0 |
| 15 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 165.0 | 151.0 | 7.0 | 74.0 | 0.0 | 7.0 | 0.0 | 6.0 |
| 16 | 12.0 | 95.0 | 41.0 | 17.0 | 26.0 | 61.0 | 30.0 | 41.0 | 3.0 | 3.0 | 0.0 | 1.0 | 5.0 | 31.0 | 36.0 | 8.0 |
| 17 | 9.0 | 17.0 | 11.0 | 9.0 | 39.0 | 59.0 | 17.0 | 75.0 | 16.0 | 24.0 | 24.0 | 43.0 | 13.0 | 18.0 | 26.0 | 10.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.133 | -0.665 | -0.347 | 0.188 | 0.25 | -0.065 | 0.389 | 0.308 | -0.294 | 0.279 | -0.742 | 0.826 | -1.133 | -0.827 | 0.681 | -0.462 |
| 02 | -1.02 | -0.525 | 0.25 | -0.827 | -0.462 | -0.15 | -0.403 | 0.052 | -1.02 | 0.465 | 0.621 | -0.196 | -1.133 | 0.122 | 1.257 | -0.665 |
| 03 | -1.782 | -0.919 | -0.196 | -1.26 | -0.15 | 0.6 | -0.593 | -0.065 | -0.294 | 0.859 | 0.809 | 0.681 | -1.576 | -0.065 | 0.25 | -0.919 |
| 04 | -2.393 | -0.294 | -0.15 | -1.26 | 0.219 | 0.809 | 0.219 | -0.244 | -0.347 | 0.535 | 0.621 | -0.462 | -0.919 | -0.106 | 0.7 | -1.26 |
| 05 | -0.827 | 0.014 | -0.462 | -1.02 | -0.106 | 0.809 | -0.244 | 0.512 | -0.106 | 0.907 | 0.014 | 0.465 | -2.041 | -0.525 | -0.665 | -0.294 |
| 06 | -0.919 | -1.406 | 0.488 | -2.393 | 0.7 | 0.156 | 0.308 | 0.579 | -0.919 | -0.106 | 0.014 | -0.462 | -0.525 | -0.525 | 0.7 | 0.156 |
| 07 | -0.196 | -0.462 | 0.087 | -0.15 | -0.024 | -0.665 | -0.593 | -0.15 | -0.462 | 0.557 | 0.579 | 0.621 | -0.294 | -0.827 | 0.557 | -0.294 |
| 08 | -0.244 | -0.065 | 0.336 | -2.939 | 0.156 | 0.415 | -0.525 | -2.939 | 0.219 | 1.037 | 0.052 | -2.939 | -0.593 | 0.842 | 0.363 | -4.236 |
| 09 | -1.406 | 1.235 | -2.939 | -4.236 | -1.02 | 1.97 | -4.236 | -2.939 | -1.782 | 1.418 | -2.393 | -2.393 | -4.236 | -2.939 | -4.236 | -2.393 |
| 10 | -0.294 | -4.236 | -4.236 | -4.236 | 2.601 | -2.041 | -0.347 | -0.742 | -2.393 | -2.939 | -4.236 | -4.236 | -1.782 | -4.236 | -2.939 | -4.236 |
| 11 | -1.26 | 2.308 | -0.106 | 1.177 | -4.236 | -1.782 | -4.236 | -4.236 | -4.236 | -0.403 | -4.236 | -2.393 | -4.236 | -1.02 | -4.236 | -2.041 |
| 12 | -4.236 | -2.939 | -1.406 | -4.236 | -2.939 | -2.393 | 2.396 | -2.041 | -2.939 | -4.236 | -0.15 | -4.236 | -2.939 | -2.939 | 1.177 | -2.041 |
| 13 | -4.236 | -4.236 | -4.236 | -2.041 | -4.236 | -4.236 | -4.236 | -1.782 | -2.041 | -4.236 | -2.041 | 2.712 | -4.236 | -4.236 | -4.236 | -1.406 |
| 14 | -4.236 | -4.236 | -2.041 | -4.236 | -4.236 | -4.236 | -4.236 | -4.236 | -4.236 | -4.236 | -2.041 | -4.236 | -4.236 | -4.236 | 2.712 | -0.665 |
| 15 | -4.236 | -4.236 | -4.236 | -4.236 | -4.236 | -4.236 | -4.236 | -4.236 | 1.85 | 1.762 | -1.26 | 1.051 | -4.236 | -1.26 | -4.236 | -1.406 |
| 16 | -0.742 | 1.3 | 0.465 | -0.403 | 0.014 | 0.859 | 0.156 | 0.465 | -2.041 | -2.041 | -4.236 | -2.939 | -1.576 | 0.188 | 0.336 | -1.133 |
| 17 | -1.02 | -0.403 | -0.827 | -1.02 | 0.415 | 0.826 | -0.403 | 1.064 | -0.462 | -0.065 | -0.065 | 0.512 | -0.665 | -0.347 | 0.014 | -0.919 |