Model info
| Transcription factor | T | ||||||||
| Model | BRAC_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 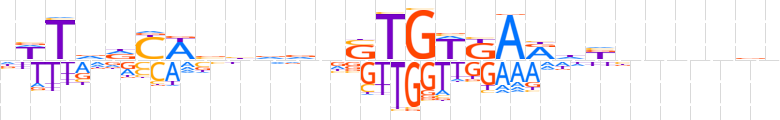 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 27 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nWTvdYWbbddnSTGWKARdbnnnndn | ||||||||
| Best auROC (human) | 0.807 | ||||||||
| Best auROC (mouse) | 0.81 | ||||||||
| Peak sets in benchmark (human) | 9 | ||||||||
| Peak sets in benchmark (mouse) | 10 | ||||||||
| Aligned words | 284 | ||||||||
| TF family | Brachyury-related factors {6.5.1} | ||||||||
| TF subfamily | T (Brachyury) {6.5.1.0.1} | ||||||||
| MGI | MGI:98472 | ||||||||
| EntrezGene | GeneID:20997 (SSTAR profile) | ||||||||
| UniProt ID | BRAC_MOUSE | ||||||||
| UniProt AC | P20293 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | T expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 4.0 | 1.0 | 15.0 | 58.0 | 5.0 | 5.0 | 3.0 | 35.0 | 11.0 | 8.0 | 14.0 | 44.0 | 19.0 | 4.0 | 5.0 | 53.0 |
| 02 | 4.0 | 0.0 | 2.0 | 33.0 | 3.0 | 3.0 | 0.0 | 12.0 | 0.0 | 6.0 | 2.0 | 29.0 | 0.0 | 5.0 | 1.0 | 184.0 |
| 03 | 2.0 | 0.0 | 4.0 | 1.0 | 6.0 | 6.0 | 0.0 | 2.0 | 1.0 | 3.0 | 0.0 | 1.0 | 126.0 | 37.0 | 63.0 | 32.0 |
| 04 | 53.0 | 4.0 | 75.0 | 3.0 | 23.0 | 6.0 | 6.0 | 11.0 | 18.0 | 1.0 | 21.0 | 27.0 | 3.0 | 2.0 | 27.0 | 4.0 |
| 05 | 3.0 | 85.0 | 3.0 | 6.0 | 2.0 | 10.0 | 1.0 | 0.0 | 8.0 | 98.0 | 6.0 | 17.0 | 5.0 | 30.0 | 8.0 | 2.0 |
| 06 | 6.0 | 0.0 | 9.0 | 3.0 | 186.0 | 5.0 | 6.0 | 26.0 | 10.0 | 2.0 | 4.0 | 2.0 | 3.0 | 1.0 | 9.0 | 12.0 |
| 07 | 25.0 | 98.0 | 51.0 | 31.0 | 3.0 | 2.0 | 0.0 | 3.0 | 6.0 | 9.0 | 12.0 | 1.0 | 9.0 | 8.0 | 15.0 | 11.0 |
| 08 | 6.0 | 14.0 | 21.0 | 2.0 | 15.0 | 28.0 | 4.0 | 70.0 | 12.0 | 23.0 | 24.0 | 19.0 | 2.0 | 25.0 | 8.0 | 11.0 |
| 09 | 11.0 | 6.0 | 11.0 | 7.0 | 47.0 | 11.0 | 13.0 | 19.0 | 13.0 | 6.0 | 17.0 | 21.0 | 51.0 | 10.0 | 28.0 | 13.0 |
| 10 | 54.0 | 8.0 | 49.0 | 11.0 | 13.0 | 5.0 | 3.0 | 12.0 | 22.0 | 12.0 | 16.0 | 19.0 | 19.0 | 5.0 | 27.0 | 9.0 |
| 11 | 39.0 | 25.0 | 37.0 | 7.0 | 10.0 | 5.0 | 1.0 | 14.0 | 28.0 | 18.0 | 32.0 | 17.0 | 6.0 | 10.0 | 15.0 | 20.0 |
| 12 | 2.0 | 13.0 | 66.0 | 2.0 | 8.0 | 33.0 | 8.0 | 9.0 | 0.0 | 8.0 | 71.0 | 6.0 | 1.0 | 12.0 | 45.0 | 0.0 |
| 13 | 0.0 | 0.0 | 0.0 | 11.0 | 0.0 | 1.0 | 0.0 | 65.0 | 4.0 | 7.0 | 2.0 | 177.0 | 1.0 | 2.0 | 1.0 | 13.0 |
| 14 | 0.0 | 1.0 | 3.0 | 1.0 | 0.0 | 0.0 | 10.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 10.0 | 255.0 | 1.0 |
| 15 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 11.0 | 34.0 | 21.0 | 12.0 | 204.0 | 1.0 | 0.0 | 0.0 | 1.0 |
| 16 | 0.0 | 8.0 | 24.0 | 3.0 | 3.0 | 3.0 | 6.0 | 9.0 | 0.0 | 2.0 | 9.0 | 1.0 | 6.0 | 16.0 | 161.0 | 33.0 |
| 17 | 8.0 | 0.0 | 1.0 | 0.0 | 27.0 | 1.0 | 1.0 | 0.0 | 189.0 | 4.0 | 6.0 | 1.0 | 41.0 | 0.0 | 1.0 | 4.0 |
| 18 | 178.0 | 15.0 | 47.0 | 25.0 | 2.0 | 1.0 | 1.0 | 1.0 | 6.0 | 2.0 | 0.0 | 1.0 | 2.0 | 0.0 | 3.0 | 0.0 |
| 19 | 82.0 | 13.0 | 25.0 | 68.0 | 5.0 | 4.0 | 0.0 | 9.0 | 14.0 | 9.0 | 14.0 | 14.0 | 6.0 | 7.0 | 7.0 | 7.0 |
| 20 | 10.0 | 18.0 | 21.0 | 58.0 | 4.0 | 1.0 | 1.0 | 27.0 | 8.0 | 5.0 | 8.0 | 25.0 | 5.0 | 15.0 | 20.0 | 58.0 |
| 21 | 3.0 | 12.0 | 9.0 | 3.0 | 12.0 | 12.0 | 3.0 | 12.0 | 21.0 | 12.0 | 8.0 | 9.0 | 19.0 | 57.0 | 42.0 | 50.0 |
| 22 | 3.0 | 20.0 | 15.0 | 17.0 | 25.0 | 44.0 | 2.0 | 22.0 | 10.0 | 16.0 | 15.0 | 21.0 | 5.0 | 24.0 | 33.0 | 12.0 |
| 23 | 9.0 | 11.0 | 15.0 | 8.0 | 13.0 | 26.0 | 10.0 | 55.0 | 12.0 | 10.0 | 27.0 | 16.0 | 10.0 | 17.0 | 21.0 | 24.0 |
| 24 | 9.0 | 13.0 | 14.0 | 8.0 | 27.0 | 11.0 | 5.0 | 21.0 | 12.0 | 28.0 | 13.0 | 20.0 | 3.0 | 13.0 | 36.0 | 51.0 |
| 25 | 11.0 | 15.0 | 22.0 | 3.0 | 25.0 | 8.0 | 16.0 | 16.0 | 15.0 | 10.0 | 26.0 | 17.0 | 29.0 | 5.0 | 49.0 | 17.0 |
| 26 | 24.0 | 30.0 | 21.0 | 5.0 | 14.0 | 10.0 | 2.0 | 12.0 | 36.0 | 31.0 | 33.0 | 13.0 | 6.0 | 7.0 | 21.0 | 19.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.425 | -2.594 | -0.165 | 1.17 | -1.218 | -1.218 | -1.686 | 0.669 | -0.467 | -0.773 | -0.232 | 0.896 | 0.067 | -1.425 | -1.218 | 1.081 |
| 02 | -1.425 | -3.937 | -2.04 | 0.611 | -1.686 | -1.686 | -3.937 | -0.382 | -3.937 | -1.047 | -2.04 | 0.483 | -3.937 | -1.218 | -2.594 | 2.321 |
| 03 | -2.04 | -3.937 | -1.425 | -2.594 | -1.047 | -1.047 | -3.937 | -2.04 | -2.594 | -1.686 | -3.937 | -2.594 | 1.943 | 0.724 | 1.253 | 0.581 |
| 04 | 1.081 | -1.425 | 1.426 | -1.686 | 0.255 | -1.047 | -1.047 | -0.467 | 0.014 | -2.594 | 0.165 | 0.413 | -1.686 | -2.04 | 0.413 | -1.425 |
| 05 | -1.686 | 1.551 | -1.686 | -1.047 | -2.04 | -0.559 | -2.594 | -3.937 | -0.773 | 1.692 | -1.047 | -0.042 | -1.218 | 0.517 | -0.773 | -2.04 |
| 06 | -1.047 | -3.937 | -0.66 | -1.686 | 2.332 | -1.218 | -1.047 | 0.376 | -0.559 | -2.04 | -1.425 | -2.04 | -1.686 | -2.594 | -0.66 | -0.382 |
| 07 | 0.337 | 1.692 | 1.043 | 0.549 | -1.686 | -2.04 | -3.937 | -1.686 | -1.047 | -0.66 | -0.382 | -2.594 | -0.66 | -0.773 | -0.165 | -0.467 |
| 08 | -1.047 | -0.232 | 0.165 | -2.04 | -0.165 | 0.449 | -1.425 | 1.357 | -0.382 | 0.255 | 0.297 | 0.067 | -2.04 | 0.337 | -0.773 | -0.467 |
| 09 | -0.467 | -1.047 | -0.467 | -0.901 | 0.962 | -0.467 | -0.304 | 0.067 | -0.304 | -1.047 | -0.042 | 0.165 | 1.043 | -0.559 | 0.449 | -0.304 |
| 10 | 1.099 | -0.773 | 1.003 | -0.467 | -0.304 | -1.218 | -1.686 | -0.382 | 0.211 | -0.382 | -0.102 | 0.067 | 0.067 | -1.218 | 0.413 | -0.66 |
| 11 | 0.776 | 0.337 | 0.724 | -0.901 | -0.559 | -1.218 | -2.594 | -0.232 | 0.449 | 0.014 | 0.581 | -0.042 | -1.047 | -0.559 | -0.165 | 0.117 |
| 12 | -2.04 | -0.304 | 1.299 | -2.04 | -0.773 | 0.611 | -0.773 | -0.66 | -3.937 | -0.773 | 1.372 | -1.047 | -2.594 | -0.382 | 0.918 | -3.937 |
| 13 | -3.937 | -3.937 | -3.937 | -0.467 | -3.937 | -2.594 | -3.937 | 1.284 | -1.425 | -0.901 | -2.04 | 2.282 | -2.594 | -2.04 | -2.594 | -0.304 |
| 14 | -3.937 | -2.594 | -1.686 | -2.594 | -3.937 | -3.937 | -0.559 | -3.937 | -3.937 | -3.937 | -1.686 | -3.937 | -3.937 | -0.559 | 2.647 | -2.594 |
| 15 | -3.937 | -3.937 | -3.937 | -3.937 | -3.937 | -3.937 | -3.937 | -0.467 | 0.641 | 0.165 | -0.382 | 2.424 | -2.594 | -3.937 | -3.937 | -2.594 |
| 16 | -3.937 | -0.773 | 0.297 | -1.686 | -1.686 | -1.686 | -1.047 | -0.66 | -3.937 | -2.04 | -0.66 | -2.594 | -1.047 | -0.102 | 2.188 | 0.611 |
| 17 | -0.773 | -3.937 | -2.594 | -3.937 | 0.413 | -2.594 | -2.594 | -3.937 | 2.348 | -1.425 | -1.047 | -2.594 | 0.826 | -3.937 | -2.594 | -1.425 |
| 18 | 2.288 | -0.165 | 0.962 | 0.337 | -2.04 | -2.594 | -2.594 | -2.594 | -1.047 | -2.04 | -3.937 | -2.594 | -2.04 | -3.937 | -1.686 | -3.937 |
| 19 | 1.515 | -0.304 | 0.337 | 1.329 | -1.218 | -1.425 | -3.937 | -0.66 | -0.232 | -0.66 | -0.232 | -0.232 | -1.047 | -0.901 | -0.901 | -0.901 |
| 20 | -0.559 | 0.014 | 0.165 | 1.17 | -1.425 | -2.594 | -2.594 | 0.413 | -0.773 | -1.218 | -0.773 | 0.337 | -1.218 | -0.165 | 0.117 | 1.17 |
| 21 | -1.686 | -0.382 | -0.66 | -1.686 | -0.382 | -0.382 | -1.686 | -0.382 | 0.165 | -0.382 | -0.773 | -0.66 | 0.067 | 1.153 | 0.85 | 1.023 |
| 22 | -1.686 | 0.117 | -0.165 | -0.042 | 0.337 | 0.896 | -2.04 | 0.211 | -0.559 | -0.102 | -0.165 | 0.165 | -1.218 | 0.297 | 0.611 | -0.382 |
| 23 | -0.66 | -0.467 | -0.165 | -0.773 | -0.304 | 0.376 | -0.559 | 1.118 | -0.382 | -0.559 | 0.413 | -0.102 | -0.559 | -0.042 | 0.165 | 0.297 |
| 24 | -0.66 | -0.304 | -0.232 | -0.773 | 0.413 | -0.467 | -1.218 | 0.165 | -0.382 | 0.449 | -0.304 | 0.117 | -1.686 | -0.304 | 0.697 | 1.043 |
| 25 | -0.467 | -0.165 | 0.211 | -1.686 | 0.337 | -0.773 | -0.102 | -0.102 | -0.165 | -0.559 | 0.376 | -0.042 | 0.483 | -1.218 | 1.003 | -0.042 |
| 26 | 0.297 | 0.517 | 0.165 | -1.218 | -0.232 | -0.559 | -2.04 | -0.382 | 0.697 | 0.549 | 0.611 | -0.304 | -1.047 | -0.901 | 0.165 | 0.067 |