Model info
| Transcription factor | CEBPA (GeneCards) | ||||||||
| Model | CEBPA_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 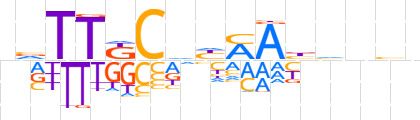 | ||||||||
| LOGO (reverse complement) | 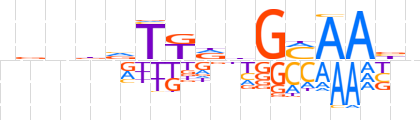 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvTTKCdhMAbhnbn | ||||||||
| Best auROC (human) | 0.988 | ||||||||
| Best auROC (mouse) | 0.991 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 153 | ||||||||
| Aligned words | 463 | ||||||||
| TF family | C/EBP-related {1.1.8} | ||||||||
| TF subfamily | C/EBP {1.1.8.1} | ||||||||
| HGNC | HGNC:1833 | ||||||||
| EntrezGene | GeneID:1050 (SSTAR profile) | ||||||||
| UniProt ID | CEBPA_HUMAN | ||||||||
| UniProt AC | P49715 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | CEBPA expression | ||||||||
| ReMap ChIP-seq dataset list | CEBPA datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 22.0 | 17.0 | 42.0 | 0.0 | 47.0 | 28.0 | 10.0 | 5.0 | 58.0 | 34.0 | 49.0 | 7.0 | 54.0 | 47.0 | 36.0 | 4.0 |
| 02 | 1.0 | 0.0 | 2.0 | 178.0 | 3.0 | 5.0 | 1.0 | 117.0 | 1.0 | 3.0 | 1.0 | 132.0 | 1.0 | 0.0 | 2.0 | 13.0 |
| 03 | 0.0 | 1.0 | 0.0 | 5.0 | 1.0 | 1.0 | 0.0 | 6.0 | 0.0 | 0.0 | 2.0 | 4.0 | 2.0 | 5.0 | 25.0 | 408.0 |
| 04 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 3.0 | 2.0 | 6.0 | 0.0 | 9.0 | 12.0 | 40.0 | 12.0 | 285.0 | 86.0 |
| 05 | 1.0 | 43.0 | 1.0 | 2.0 | 0.0 | 13.0 | 0.0 | 1.0 | 1.0 | 291.0 | 5.0 | 1.0 | 1.0 | 97.0 | 1.0 | 2.0 |
| 06 | 1.0 | 1.0 | 1.0 | 0.0 | 188.0 | 68.0 | 108.0 | 80.0 | 2.0 | 3.0 | 2.0 | 0.0 | 1.0 | 2.0 | 0.0 | 3.0 |
| 07 | 45.0 | 98.0 | 7.0 | 42.0 | 16.0 | 36.0 | 2.0 | 20.0 | 15.0 | 43.0 | 0.0 | 53.0 | 9.0 | 51.0 | 3.0 | 20.0 |
| 08 | 42.0 | 40.0 | 0.0 | 3.0 | 173.0 | 51.0 | 1.0 | 3.0 | 5.0 | 4.0 | 0.0 | 3.0 | 45.0 | 87.0 | 1.0 | 2.0 |
| 09 | 226.0 | 10.0 | 17.0 | 12.0 | 174.0 | 1.0 | 5.0 | 2.0 | 0.0 | 2.0 | 0.0 | 0.0 | 6.0 | 2.0 | 1.0 | 2.0 |
| 10 | 16.0 | 169.0 | 67.0 | 154.0 | 6.0 | 4.0 | 0.0 | 5.0 | 7.0 | 4.0 | 7.0 | 5.0 | 4.0 | 2.0 | 0.0 | 10.0 |
| 11 | 12.0 | 13.0 | 3.0 | 5.0 | 64.0 | 64.0 | 10.0 | 41.0 | 23.0 | 26.0 | 11.0 | 14.0 | 30.0 | 70.0 | 23.0 | 51.0 |
| 12 | 23.0 | 39.0 | 47.0 | 20.0 | 49.0 | 67.0 | 18.0 | 39.0 | 14.0 | 13.0 | 8.0 | 12.0 | 26.0 | 33.0 | 17.0 | 35.0 |
| 13 | 13.0 | 44.0 | 38.0 | 17.0 | 23.0 | 69.0 | 15.0 | 45.0 | 13.0 | 34.0 | 31.0 | 12.0 | 11.0 | 54.0 | 23.0 | 18.0 |
| 14 | 10.0 | 18.0 | 23.0 | 9.0 | 55.0 | 83.0 | 18.0 | 45.0 | 7.0 | 49.0 | 29.0 | 22.0 | 9.0 | 16.0 | 30.0 | 37.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.264 | -0.516 | 0.375 | -4.331 | 0.486 | -0.026 | -1.032 | -1.689 | 0.695 | 0.166 | 0.528 | -1.373 | 0.624 | 0.486 | 0.222 | -1.894 |
| 02 | -3.047 | -4.331 | -2.503 | 1.812 | -2.153 | -1.689 | -3.047 | 1.394 | -3.047 | -2.153 | -3.047 | 1.514 | -3.047 | -4.331 | -2.503 | -0.778 |
| 03 | -4.331 | -3.047 | -4.331 | -1.689 | -3.047 | -3.047 | -4.331 | -1.518 | -4.331 | -4.331 | -2.503 | -1.894 | -2.503 | -1.689 | -0.138 | 2.64 |
| 04 | -4.331 | -3.047 | -3.047 | -3.047 | -3.047 | -3.047 | -2.153 | -2.503 | -1.518 | -4.331 | -1.133 | -0.856 | 0.327 | -0.856 | 2.282 | 1.087 |
| 05 | -3.047 | 0.398 | -3.047 | -2.503 | -4.331 | -0.778 | -4.331 | -3.047 | -3.047 | 2.303 | -1.689 | -3.047 | -3.047 | 1.207 | -3.047 | -2.503 |
| 06 | -3.047 | -3.047 | -3.047 | -4.331 | 1.867 | 0.853 | 1.314 | 1.015 | -2.503 | -2.153 | -2.503 | -4.331 | -3.047 | -2.503 | -4.331 | -2.153 |
| 07 | 0.443 | 1.217 | -1.373 | 0.375 | -0.576 | 0.222 | -2.503 | -0.357 | -0.639 | 0.398 | -4.331 | 0.606 | -1.133 | 0.567 | -2.153 | -0.357 |
| 08 | 0.375 | 0.327 | -4.331 | -2.153 | 1.784 | 0.567 | -3.047 | -2.153 | -1.689 | -1.894 | -4.331 | -2.153 | 0.443 | 1.098 | -3.047 | -2.503 |
| 09 | 2.05 | -1.032 | -0.516 | -0.856 | 1.789 | -3.047 | -1.689 | -2.503 | -4.331 | -2.503 | -4.331 | -4.331 | -1.518 | -2.503 | -3.047 | -2.503 |
| 10 | -0.576 | 1.76 | 0.839 | 1.668 | -1.518 | -1.894 | -4.331 | -1.689 | -1.373 | -1.894 | -1.373 | -1.689 | -1.894 | -2.503 | -4.331 | -1.032 |
| 11 | -0.856 | -0.778 | -2.153 | -1.689 | 0.793 | 0.793 | -1.032 | 0.351 | -0.22 | -0.099 | -0.94 | -0.706 | 0.042 | 0.882 | -0.22 | 0.567 |
| 12 | -0.22 | 0.301 | 0.486 | -0.357 | 0.528 | 0.839 | -0.46 | 0.301 | -0.706 | -0.778 | -1.246 | -0.856 | -0.099 | 0.136 | -0.516 | 0.194 |
| 13 | -0.778 | 0.421 | 0.276 | -0.516 | -0.22 | 0.868 | -0.639 | 0.443 | -0.778 | 0.166 | 0.074 | -0.856 | -0.94 | 0.624 | -0.22 | -0.46 |
| 14 | -1.032 | -0.46 | -0.22 | -1.133 | 0.642 | 1.052 | -0.46 | 0.443 | -1.373 | 0.528 | 0.009 | -0.264 | -1.133 | -0.576 | 0.042 | 0.249 |