Model info
| Transcription factor | Ebf1 | ||||||||
| Model | COE1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 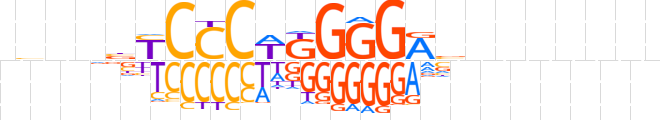 | ||||||||
| LOGO (reverse complement) | 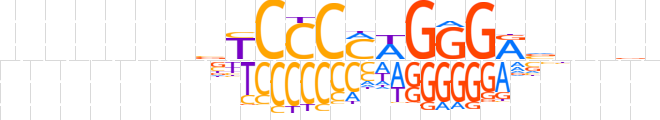 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvndbYCCCWKGGGRvhnnnnnn | ||||||||
| Best auROC (human) | 0.996 | ||||||||
| Best auROC (mouse) | 0.994 | ||||||||
| Peak sets in benchmark (human) | 17 | ||||||||
| Peak sets in benchmark (mouse) | 55 | ||||||||
| Aligned words | 506 | ||||||||
| TF family | Early B-Cell Factor-related factors {6.1.5} | ||||||||
| TF subfamily | EBF1 (COE1) {6.1.5.0.1} | ||||||||
| MGI | MGI:95275 | ||||||||
| EntrezGene | GeneID:13591 (SSTAR profile) | ||||||||
| UniProt ID | COE1_MOUSE | ||||||||
| UniProt AC | Q07802 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Ebf1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 20.07 | 20.922 | 39.115 | 7.54 | 38.651 | 38.664 | 11.701 | 15.4 | 18.325 | 63.547 | 20.493 | 31.719 | 6.102 | 60.734 | 39.853 | 17.677 |
| 02 | 20.4 | 18.914 | 39.954 | 3.881 | 60.731 | 62.927 | 16.514 | 43.695 | 18.623 | 33.14 | 31.756 | 27.642 | 6.336 | 18.084 | 29.173 | 18.742 |
| 03 | 36.237 | 11.382 | 47.689 | 10.781 | 60.659 | 28.274 | 9.336 | 34.797 | 22.417 | 16.849 | 55.118 | 23.013 | 24.767 | 10.44 | 43.304 | 15.449 |
| 04 | 17.75 | 16.734 | 76.568 | 33.029 | 12.572 | 37.048 | 7.64 | 9.685 | 9.285 | 46.27 | 72.565 | 27.327 | 2.133 | 31.83 | 41.684 | 8.394 |
| 05 | 0.0 | 3.72 | 2.097 | 35.923 | 12.394 | 9.535 | 0.0 | 109.954 | 5.793 | 53.795 | 7.429 | 131.441 | 4.214 | 12.75 | 5.227 | 56.243 |
| 06 | 0.0 | 22.401 | 0.0 | 0.0 | 0.0 | 78.621 | 0.0 | 1.179 | 0.0 | 14.752 | 0.0 | 0.0 | 0.0 | 329.877 | 3.683 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 4.237 | 371.708 | 0.0 | 69.706 | 0.0 | 1.7 | 0.0 | 1.983 | 0.0 | 1.179 | 0.0 | 0.0 |
| 08 | 0.0 | 4.237 | 0.0 | 0.0 | 1.116 | 372.583 | 0.0 | 0.889 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 71.69 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 1.116 | 164.282 | 4.033 | 11.983 | 268.212 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.889 |
| 10 | 0.0 | 0.0 | 131.792 | 32.49 | 0.0 | 0.0 | 4.033 | 0.0 | 0.858 | 0.0 | 11.125 | 0.0 | 28.797 | 1.982 | 178.699 | 60.738 |
| 11 | 0.0 | 0.0 | 29.655 | 0.0 | 0.936 | 0.0 | 1.046 | 0.0 | 2.198 | 0.906 | 320.951 | 1.594 | 0.0 | 0.0 | 93.228 | 0.0 |
| 12 | 0.0 | 0.0 | 3.134 | 0.0 | 0.0 | 0.0 | 0.906 | 0.0 | 63.956 | 0.0 | 380.923 | 0.0 | 0.0 | 0.0 | 1.594 | 0.0 |
| 13 | 0.0 | 0.0 | 63.956 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 386.558 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 333.692 | 13.451 | 95.871 | 7.5 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 56.22 | 135.865 | 104.535 | 37.073 | 5.926 | 3.802 | 0.0 | 3.723 | 33.611 | 45.963 | 9.731 | 6.565 | 0.0 | 3.627 | 2.052 | 1.821 |
| 16 | 18.125 | 25.976 | 11.374 | 40.283 | 57.452 | 83.214 | 7.507 | 41.084 | 23.023 | 36.658 | 27.87 | 28.767 | 5.48 | 12.868 | 7.948 | 22.885 |
| 17 | 19.761 | 30.787 | 26.053 | 27.479 | 46.732 | 44.687 | 10.207 | 57.091 | 14.435 | 9.768 | 17.637 | 12.859 | 19.84 | 31.427 | 23.903 | 57.848 |
| 18 | 24.738 | 19.177 | 46.557 | 10.296 | 35.796 | 30.804 | 11.115 | 38.954 | 18.279 | 22.879 | 25.233 | 11.409 | 21.253 | 39.017 | 75.136 | 19.871 |
| 19 | 21.565 | 13.674 | 56.411 | 8.417 | 35.24 | 23.901 | 8.955 | 43.782 | 38.329 | 41.569 | 46.596 | 31.547 | 6.524 | 12.736 | 42.366 | 18.903 |
| 20 | 11.861 | 29.654 | 51.374 | 8.77 | 34.078 | 24.524 | 7.356 | 25.921 | 29.77 | 38.259 | 65.767 | 20.531 | 25.553 | 27.669 | 36.631 | 12.795 |
| 21 | 16.74 | 14.562 | 53.974 | 15.985 | 40.975 | 38.529 | 11.272 | 29.33 | 29.082 | 23.933 | 73.356 | 34.757 | 8.035 | 20.055 | 30.077 | 9.851 |
| 22 | 18.615 | 27.521 | 33.701 | 14.995 | 23.566 | 33.833 | 4.83 | 34.851 | 30.807 | 26.488 | 76.363 | 35.022 | 7.463 | 26.947 | 32.604 | 22.909 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.333 | -0.292 | 0.325 | -1.282 | 0.313 | 0.313 | -0.859 | -0.592 | -0.422 | 0.807 | -0.313 | 0.118 | -1.482 | 0.761 | 0.343 | -0.458 |
| 02 | -0.317 | -0.391 | 0.346 | -1.901 | 0.761 | 0.797 | -0.524 | 0.435 | -0.407 | 0.161 | 0.119 | -0.018 | -1.446 | -0.435 | 0.035 | -0.4 |
| 03 | 0.249 | -0.886 | 0.521 | -0.939 | 0.76 | 0.004 | -1.077 | 0.209 | -0.225 | -0.505 | 0.665 | -0.199 | -0.126 | -0.97 | 0.426 | -0.589 |
| 04 | -0.454 | -0.511 | 0.992 | 0.158 | -0.79 | 0.271 | -1.269 | -1.042 | -1.083 | 0.491 | 0.938 | -0.03 | -2.429 | 0.121 | 0.388 | -1.179 |
| 05 | -4.314 | -1.94 | -2.444 | 0.241 | -0.804 | -1.057 | -4.314 | 1.352 | -1.531 | 0.641 | -1.296 | 1.53 | -1.826 | -0.776 | -1.627 | 0.685 |
| 06 | -4.314 | -0.225 | -4.314 | -4.314 | -4.314 | 1.018 | -4.314 | -2.906 | -4.314 | -0.634 | -4.314 | -4.314 | -4.314 | 2.449 | -1.949 | -4.314 |
| 07 | -4.314 | -4.314 | -4.314 | -4.314 | -1.821 | 2.568 | -4.314 | 0.898 | -4.314 | -2.618 | -4.314 | -2.49 | -4.314 | -2.906 | -4.314 | -4.314 |
| 08 | -4.314 | -1.821 | -4.314 | -4.314 | -2.947 | 2.57 | -4.314 | -3.112 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | 0.926 | -4.314 | -4.314 |
| 09 | -4.314 | -4.314 | -4.314 | -2.947 | 1.753 | -1.866 | -0.836 | 2.242 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | -3.112 |
| 10 | -4.314 | -4.314 | 1.533 | 0.141 | -4.314 | -4.314 | -1.866 | -4.314 | -3.137 | -4.314 | -0.908 | -4.314 | 0.022 | -2.491 | 1.837 | 0.762 |
| 11 | -4.314 | -4.314 | 0.051 | -4.314 | -3.075 | -4.314 | -2.995 | -4.314 | -2.403 | -3.099 | 2.421 | -2.67 | -4.314 | -4.314 | 1.188 | -4.314 |
| 12 | -4.314 | -4.314 | -2.094 | -4.314 | -4.314 | -4.314 | -3.099 | -4.314 | 0.813 | -4.314 | 2.592 | -4.314 | -4.314 | -4.314 | -2.67 | -4.314 |
| 13 | -4.314 | -4.314 | 0.813 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | 2.607 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 |
| 14 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | -4.314 | 2.46 | -0.724 | 1.216 | -1.287 | -4.314 | -4.314 | -4.314 | -4.314 |
| 15 | 0.685 | 1.563 | 1.302 | 0.272 | -1.509 | -1.92 | -4.314 | -1.939 | 0.175 | 0.485 | -1.037 | -1.413 | -4.314 | -1.963 | -2.462 | -2.562 |
| 16 | -0.433 | -0.08 | -0.887 | 0.354 | 0.706 | 1.075 | -1.286 | 0.374 | -0.198 | 0.261 | -0.01 | 0.021 | -1.583 | -0.767 | -1.231 | -0.204 |
| 17 | -0.348 | 0.088 | -0.077 | -0.024 | 0.501 | 0.457 | -0.991 | 0.7 | -0.655 | -1.034 | -0.46 | -0.768 | -0.344 | 0.108 | -0.161 | 0.713 |
| 18 | -0.128 | -0.378 | 0.498 | -0.983 | 0.237 | 0.089 | -0.909 | 0.321 | -0.425 | -0.204 | -0.108 | -0.884 | -0.277 | 0.322 | 0.973 | -0.343 |
| 19 | -0.263 | -0.708 | 0.688 | -1.177 | 0.222 | -0.162 | -1.117 | 0.437 | 0.305 | 0.385 | 0.498 | 0.112 | -1.419 | -0.777 | 0.404 | -0.392 |
| 20 | -0.846 | 0.051 | 0.595 | -1.137 | 0.189 | -0.136 | -1.305 | -0.082 | 0.055 | 0.303 | 0.841 | -0.311 | -0.096 | -0.017 | 0.26 | -0.773 |
| 21 | -0.511 | -0.647 | 0.644 | -0.556 | 0.371 | 0.31 | -0.896 | 0.04 | 0.032 | -0.16 | 0.949 | 0.208 | -1.221 | -0.334 | 0.065 | -1.026 |
| 22 | -0.407 | -0.023 | 0.178 | -0.618 | -0.175 | 0.181 | -1.7 | 0.211 | 0.089 | -0.06 | 0.989 | 0.216 | -1.291 | -0.043 | 0.145 | -0.203 |