Model info
| Transcription factor | Creb1 | ||||||||
| Model | CREB1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 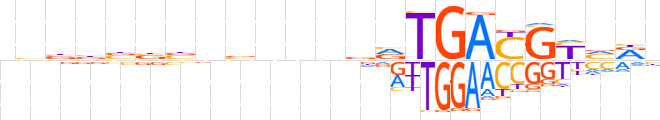 | ||||||||
| LOGO (reverse complement) | 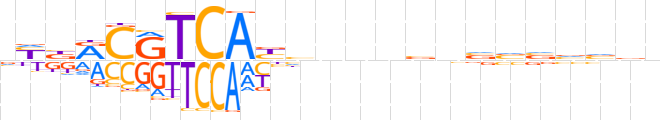 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nhbbbbbnbnnndRTGAYGYvRn | ||||||||
| Best auROC (human) | 0.954 | ||||||||
| Best auROC (mouse) | 0.907 | ||||||||
| Peak sets in benchmark (human) | 63 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 508 | ||||||||
| TF family | CREB-related factors {1.1.7} | ||||||||
| TF subfamily | CREB-like factors {1.1.7.1} | ||||||||
| MGI | MGI:88494 | ||||||||
| EntrezGene | GeneID:12912 (SSTAR profile) | ||||||||
| UniProt ID | CREB1_MOUSE | ||||||||
| UniProt AC | Q01147 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Creb1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 28.434 | 20.34 | 18.042 | 22.52 | 24.882 | 75.834 | 9.568 | 19.801 | 25.035 | 69.217 | 26.068 | 32.705 | 9.885 | 44.168 | 30.758 | 42.743 |
| 02 | 8.198 | 19.729 | 54.273 | 6.037 | 32.294 | 44.295 | 94.647 | 38.323 | 12.642 | 19.273 | 32.365 | 20.156 | 8.676 | 32.649 | 58.929 | 17.515 |
| 03 | 12.317 | 16.467 | 20.207 | 12.819 | 16.4 | 34.612 | 23.585 | 41.348 | 14.596 | 43.043 | 130.532 | 52.043 | 6.86 | 20.583 | 28.641 | 25.946 |
| 04 | 5.248 | 19.244 | 16.003 | 9.677 | 20.056 | 39.941 | 34.089 | 20.619 | 14.019 | 132.526 | 32.591 | 23.831 | 0.0 | 62.352 | 38.99 | 30.814 |
| 05 | 12.172 | 3.477 | 17.771 | 5.902 | 23.364 | 38.853 | 171.693 | 20.153 | 16.559 | 41.268 | 41.325 | 22.521 | 11.927 | 14.128 | 27.286 | 31.6 |
| 06 | 13.747 | 18.676 | 26.87 | 4.729 | 19.319 | 37.015 | 28.626 | 12.766 | 15.239 | 177.716 | 41.002 | 24.118 | 9.902 | 23.188 | 29.558 | 17.529 |
| 07 | 11.421 | 15.701 | 21.7 | 9.386 | 76.602 | 51.256 | 89.023 | 39.714 | 16.639 | 44.877 | 50.015 | 14.525 | 2.574 | 16.967 | 28.214 | 11.387 |
| 08 | 16.193 | 29.483 | 42.216 | 19.342 | 19.566 | 53.298 | 27.28 | 28.657 | 26.315 | 88.59 | 51.79 | 22.257 | 3.91 | 35.487 | 17.655 | 17.961 |
| 09 | 16.379 | 12.404 | 29.71 | 7.491 | 49.636 | 62.08 | 56.812 | 38.331 | 33.333 | 38.301 | 43.098 | 24.21 | 11.759 | 23.918 | 39.975 | 12.564 |
| 10 | 24.517 | 32.88 | 40.964 | 12.746 | 34.922 | 42.045 | 24.123 | 35.614 | 23.517 | 58.075 | 62.991 | 25.011 | 15.525 | 12.756 | 22.929 | 31.386 |
| 11 | 23.115 | 27.492 | 26.439 | 21.435 | 19.024 | 64.354 | 9.469 | 52.908 | 36.832 | 50.443 | 32.083 | 31.649 | 9.592 | 30.518 | 26.591 | 38.056 |
| 12 | 40.363 | 9.257 | 29.859 | 9.084 | 39.397 | 26.897 | 58.03 | 48.483 | 15.714 | 9.471 | 35.81 | 33.587 | 24.71 | 9.577 | 76.776 | 32.984 |
| 13 | 53.042 | 7.612 | 54.968 | 4.563 | 21.233 | 7.527 | 24.396 | 2.046 | 102.737 | 32.717 | 62.967 | 2.054 | 29.516 | 23.07 | 69.028 | 2.524 |
| 14 | 3.841 | 5.343 | 4.066 | 193.279 | 1.072 | 4.712 | 1.09 | 64.053 | 1.881 | 6.247 | 0.0 | 203.232 | 0.0 | 0.0 | 0.0 | 11.187 |
| 15 | 0.0 | 0.0 | 6.793 | 0.0 | 0.0 | 0.947 | 14.555 | 0.799 | 0.0 | 0.788 | 4.368 | 0.0 | 3.472 | 0.0 | 462.166 | 6.111 |
| 16 | 3.472 | 0.0 | 0.0 | 0.0 | 0.788 | 0.0 | 0.947 | 0.0 | 453.939 | 1.688 | 26.44 | 5.816 | 5.806 | 1.104 | 0.0 | 0.0 |
| 17 | 2.488 | 347.69 | 27.36 | 86.467 | 0.0 | 1.971 | 0.0 | 0.821 | 1.104 | 23.902 | 0.0 | 2.38 | 0.0 | 5.816 | 0.0 | 0.0 |
| 18 | 0.0 | 0.0 | 3.592 | 0.0 | 22.046 | 21.657 | 329.813 | 5.864 | 1.552 | 2.358 | 20.899 | 2.552 | 0.849 | 6.594 | 78.899 | 3.325 |
| 19 | 0.0 | 3.509 | 5.835 | 15.103 | 3.46 | 2.673 | 1.094 | 23.382 | 36.677 | 91.685 | 15.499 | 289.342 | 2.269 | 0.776 | 0.0 | 8.696 |
| 20 | 10.378 | 23.984 | 3.784 | 4.261 | 60.15 | 15.301 | 11.203 | 11.99 | 10.199 | 8.229 | 2.906 | 1.094 | 56.127 | 214.167 | 35.691 | 30.537 |
| 21 | 66.196 | 19.465 | 39.972 | 11.22 | 213.451 | 9.826 | 10.873 | 27.531 | 21.177 | 8.333 | 17.25 | 6.824 | 14.544 | 1.161 | 20.859 | 11.317 |
| 22 | 53.326 | 98.144 | 78.767 | 85.132 | 12.899 | 9.922 | 11.522 | 4.442 | 15.279 | 38.559 | 25.108 | 10.008 | 9.162 | 21.712 | 11.073 | 14.945 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.093 | -0.423 | -0.54 | -0.323 | -0.225 | 0.879 | -1.156 | -0.449 | -0.219 | 0.788 | -0.179 | 0.045 | -1.125 | 0.342 | -0.016 | 0.31 |
| 02 | -1.304 | -0.453 | 0.547 | -1.594 | 0.032 | 0.345 | 1.1 | 0.202 | -0.887 | -0.476 | 0.035 | -0.432 | -1.25 | 0.043 | 0.629 | -0.569 |
| 03 | -0.912 | -0.63 | -0.429 | -0.874 | -0.634 | 0.101 | -0.277 | 0.277 | -0.747 | 0.317 | 1.42 | 0.505 | -1.474 | -0.411 | -0.086 | -0.183 |
| 04 | -1.725 | -0.477 | -0.658 | -1.145 | -0.437 | 0.243 | 0.086 | -0.409 | -0.787 | 1.435 | 0.041 | -0.267 | -4.4 | 0.685 | 0.219 | -0.014 |
| 05 | -0.924 | -2.102 | -0.555 | -1.615 | -0.287 | 0.215 | 1.694 | -0.432 | -0.624 | 0.275 | 0.276 | -0.323 | -0.944 | -0.779 | -0.134 | 0.011 |
| 06 | -0.806 | -0.507 | -0.149 | -1.822 | -0.473 | 0.167 | -0.087 | -0.878 | -0.705 | 1.728 | 0.269 | -0.255 | -1.123 | -0.294 | -0.055 | -0.569 |
| 07 | -0.985 | -0.676 | -0.359 | -1.175 | 0.889 | 0.49 | 1.039 | 0.237 | -0.62 | 0.358 | 0.466 | -0.752 | -2.368 | -0.6 | -0.101 | -0.988 |
| 08 | -0.646 | -0.057 | 0.298 | -0.472 | -0.461 | 0.529 | -0.134 | -0.085 | -0.17 | 1.034 | 0.5 | -0.334 | -1.996 | 0.126 | -0.562 | -0.545 |
| 09 | -0.635 | -0.906 | -0.05 | -1.39 | 0.458 | 0.68 | 0.592 | 0.202 | 0.064 | 0.201 | 0.318 | -0.252 | -0.957 | -0.264 | 0.244 | -0.893 |
| 10 | -0.239 | 0.05 | 0.268 | -0.879 | 0.11 | 0.294 | -0.255 | 0.129 | -0.28 | 0.614 | 0.695 | -0.22 | -0.687 | -0.878 | -0.305 | 0.004 |
| 11 | -0.297 | -0.126 | -0.165 | -0.371 | -0.488 | 0.716 | -1.166 | 0.521 | 0.162 | 0.474 | 0.026 | 0.013 | -1.154 | -0.023 | -0.159 | 0.195 |
| 12 | 0.253 | -1.188 | -0.045 | -1.206 | 0.229 | -0.148 | 0.613 | 0.435 | -0.675 | -1.166 | 0.135 | 0.071 | -0.232 | -1.155 | 0.892 | 0.053 |
| 13 | 0.524 | -1.375 | 0.559 | -1.855 | -0.381 | -1.386 | -0.244 | -2.565 | 1.182 | 0.045 | 0.694 | -2.561 | -0.056 | -0.299 | 0.786 | -2.385 |
| 14 | -2.012 | -1.708 | -1.961 | 1.812 | -3.076 | -1.825 | -3.063 | 0.711 | -2.635 | -1.562 | -4.4 | 1.862 | -4.4 | -4.4 | -4.4 | -1.006 |
| 15 | -4.4 | -4.4 | -1.483 | -4.4 | -4.4 | -3.165 | -0.75 | -3.282 | -4.4 | -3.292 | -1.895 | -4.4 | -2.103 | -4.4 | 2.682 | -1.583 |
| 16 | -2.103 | -4.4 | -4.4 | -4.4 | -3.292 | -4.4 | -3.165 | -4.4 | 2.664 | -2.724 | -0.165 | -1.629 | -1.631 | -3.054 | -4.4 | -4.4 |
| 17 | -2.398 | 2.398 | -0.131 | 1.01 | -4.4 | -2.596 | -4.4 | -3.265 | -3.054 | -0.264 | -4.4 | -2.436 | -4.4 | -1.629 | -4.4 | -4.4 |
| 18 | -4.4 | -4.4 | -2.073 | -4.4 | -0.344 | -0.361 | 2.345 | -1.621 | -2.792 | -2.444 | -0.396 | -2.376 | -3.241 | -1.511 | 0.919 | -2.142 |
| 19 | -4.4 | -2.094 | -1.626 | -0.714 | -2.107 | -2.335 | -3.061 | -0.286 | 0.158 | 1.068 | -0.689 | 2.215 | -2.477 | -3.302 | -4.4 | -1.248 |
| 20 | -1.078 | -0.261 | -2.026 | -1.918 | 0.649 | -0.701 | -1.004 | -0.938 | -1.095 | -1.301 | -2.262 | -3.061 | 0.58 | 1.914 | 0.131 | -0.023 |
| 21 | 0.744 | -0.466 | 0.243 | -1.003 | 1.911 | -1.131 | -1.033 | -0.125 | -0.383 | -1.289 | -0.584 | -1.479 | -0.751 | -3.017 | -0.398 | -0.994 |
| 22 | 0.529 | 1.136 | 0.917 | 0.994 | -0.868 | -1.121 | -0.977 | -1.879 | -0.703 | 0.208 | -0.216 | -1.113 | -1.198 | -0.359 | -1.015 | -0.724 |