Model info
| Transcription factor | Ctcfl | ||||||||
| Model | CTCFL_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 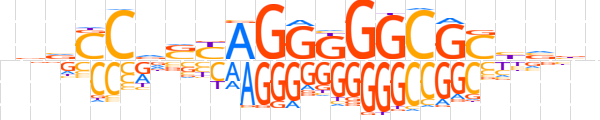 | ||||||||
| LOGO (reverse complement) | 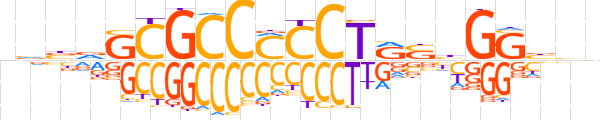 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bvvCCdSYAGGKGGCGChvbn | ||||||||
| Best auROC (human) | 0.978 | ||||||||
| Best auROC (mouse) | 0.997 | ||||||||
| Peak sets in benchmark (human) | 25 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | CTCF-like factors {2.3.3.50} | ||||||||
| MGI | MGI:3652571 | ||||||||
| EntrezGene | GeneID:664799 (SSTAR profile) | ||||||||
| UniProt ID | CTCFL_MOUSE | ||||||||
| UniProt AC | A2APF3 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Ctcfl expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 8.0 | 7.0 | 22.0 | 2.0 | 38.0 | 60.0 | 72.0 | 47.0 | 38.0 | 38.0 | 44.0 | 10.0 | 7.0 | 25.0 | 64.0 | 9.0 |
| 02 | 13.0 | 14.0 | 53.0 | 11.0 | 35.0 | 13.0 | 58.0 | 24.0 | 30.0 | 34.0 | 116.0 | 22.0 | 9.0 | 22.0 | 29.0 | 8.0 |
| 03 | 6.0 | 71.0 | 8.0 | 2.0 | 8.0 | 58.0 | 7.0 | 10.0 | 9.0 | 214.0 | 22.0 | 11.0 | 0.0 | 60.0 | 3.0 | 2.0 |
| 04 | 2.0 | 20.0 | 0.0 | 1.0 | 8.0 | 390.0 | 2.0 | 3.0 | 1.0 | 37.0 | 1.0 | 1.0 | 0.0 | 23.0 | 2.0 | 0.0 |
| 05 | 2.0 | 2.0 | 5.0 | 2.0 | 155.0 | 46.0 | 198.0 | 71.0 | 0.0 | 3.0 | 1.0 | 1.0 | 0.0 | 2.0 | 3.0 | 0.0 |
| 06 | 7.0 | 82.0 | 66.0 | 2.0 | 12.0 | 16.0 | 20.0 | 5.0 | 20.0 | 121.0 | 62.0 | 4.0 | 5.0 | 26.0 | 42.0 | 1.0 |
| 07 | 2.0 | 28.0 | 7.0 | 7.0 | 9.0 | 140.0 | 17.0 | 79.0 | 7.0 | 133.0 | 4.0 | 46.0 | 0.0 | 9.0 | 1.0 | 2.0 |
| 08 | 8.0 | 5.0 | 4.0 | 1.0 | 291.0 | 5.0 | 7.0 | 7.0 | 13.0 | 6.0 | 9.0 | 1.0 | 125.0 | 0.0 | 8.0 | 1.0 |
| 09 | 2.0 | 0.0 | 435.0 | 0.0 | 0.0 | 1.0 | 15.0 | 0.0 | 0.0 | 4.0 | 24.0 | 0.0 | 1.0 | 0.0 | 9.0 | 0.0 |
| 10 | 0.0 | 0.0 | 3.0 | 0.0 | 1.0 | 0.0 | 4.0 | 0.0 | 75.0 | 1.0 | 405.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 2.0 | 0.0 | 70.0 | 4.0 | 0.0 | 1.0 | 0.0 | 0.0 | 21.0 | 28.0 | 321.0 | 42.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 12 | 0.0 | 0.0 | 23.0 | 0.0 | 0.0 | 0.0 | 30.0 | 0.0 | 0.0 | 0.0 | 392.0 | 0.0 | 0.0 | 0.0 | 46.0 | 0.0 |
| 13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 6.0 | 458.0 | 25.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 5.0 | 1.0 | 0.0 | 19.0 | 437.0 | 1.0 | 1.0 | 0.0 | 25.0 | 0.0 | 0.0 |
| 15 | 2.0 | 0.0 | 18.0 | 0.0 | 68.0 | 1.0 | 397.0 | 2.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 16 | 0.0 | 7.0 | 63.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 15.0 | 378.0 | 13.0 | 12.0 | 0.0 | 2.0 | 0.0 | 0.0 |
| 17 | 1.0 | 9.0 | 1.0 | 4.0 | 37.0 | 130.0 | 30.0 | 190.0 | 10.0 | 36.0 | 9.0 | 21.0 | 1.0 | 6.0 | 3.0 | 3.0 |
| 18 | 3.0 | 16.0 | 28.0 | 2.0 | 72.0 | 59.0 | 37.0 | 13.0 | 5.0 | 32.0 | 5.0 | 1.0 | 16.0 | 39.0 | 153.0 | 10.0 |
| 19 | 12.0 | 31.0 | 40.0 | 13.0 | 13.0 | 31.0 | 48.0 | 54.0 | 14.0 | 96.0 | 27.0 | 86.0 | 4.0 | 9.0 | 9.0 | 4.0 |
| 20 | 13.0 | 8.0 | 17.0 | 5.0 | 51.0 | 28.0 | 50.0 | 38.0 | 42.0 | 40.0 | 27.0 | 15.0 | 22.0 | 34.0 | 73.0 | 28.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.31 | -1.437 | -0.328 | -2.566 | 0.211 | 0.664 | 0.846 | 0.422 | 0.211 | 0.211 | 0.357 | -1.096 | -1.437 | -0.202 | 0.729 | -1.197 |
| 02 | -0.842 | -0.77 | 0.541 | -1.004 | 0.13 | -0.842 | 0.631 | -0.242 | -0.022 | 0.101 | 1.321 | -0.328 | -1.197 | -0.328 | -0.056 | -1.31 |
| 03 | -1.582 | 0.832 | -1.31 | -2.566 | -1.31 | 0.631 | -1.437 | -1.096 | -1.197 | 1.931 | -0.328 | -1.004 | -4.385 | 0.664 | -2.216 | -2.566 |
| 04 | -2.566 | -0.421 | -4.385 | -3.109 | -1.31 | 2.531 | -2.566 | -2.216 | -3.109 | 0.185 | -3.109 | -3.109 | -4.385 | -0.284 | -2.566 | -4.385 |
| 05 | -2.566 | -2.566 | -1.752 | -2.566 | 1.61 | 0.401 | 1.854 | 0.832 | -4.385 | -2.216 | -3.109 | -3.109 | -4.385 | -2.566 | -2.216 | -4.385 |
| 06 | -1.437 | 0.975 | 0.759 | -2.566 | -0.92 | -0.64 | -0.421 | -1.752 | -0.421 | 1.363 | 0.697 | -1.958 | -1.752 | -0.164 | 0.31 | -3.109 |
| 07 | -2.566 | -0.09 | -1.437 | -1.437 | -1.197 | 1.508 | -0.581 | 0.938 | -1.437 | 1.457 | -1.958 | 0.401 | -4.385 | -1.197 | -3.109 | -2.566 |
| 08 | -1.31 | -1.752 | -1.958 | -3.109 | 2.238 | -1.752 | -1.437 | -1.437 | -0.842 | -1.582 | -1.197 | -3.109 | 1.395 | -4.385 | -1.31 | -3.109 |
| 09 | -2.566 | -4.385 | 2.64 | -4.385 | -4.385 | -3.109 | -0.703 | -4.385 | -4.385 | -1.958 | -0.242 | -4.385 | -3.109 | -4.385 | -1.197 | -4.385 |
| 10 | -4.385 | -4.385 | -2.216 | -4.385 | -3.109 | -4.385 | -1.958 | -4.385 | 0.886 | -3.109 | 2.568 | -2.566 | -4.385 | -4.385 | -4.385 | -4.385 |
| 11 | -2.566 | -4.385 | 0.818 | -1.958 | -4.385 | -3.109 | -4.385 | -4.385 | -0.374 | -0.09 | 2.336 | 0.31 | -4.385 | -3.109 | -3.109 | -4.385 |
| 12 | -4.385 | -4.385 | -0.284 | -4.385 | -4.385 | -4.385 | -0.022 | -4.385 | -4.385 | -4.385 | 2.536 | -4.385 | -4.385 | -4.385 | 0.401 | -4.385 |
| 13 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -2.566 | -1.582 | 2.691 | -0.202 | -4.385 | -4.385 | -4.385 | -4.385 |
| 14 | -3.109 | -3.109 | -4.385 | -4.385 | -4.385 | -1.752 | -3.109 | -4.385 | -0.472 | 2.644 | -3.109 | -3.109 | -4.385 | -0.202 | -4.385 | -4.385 |
| 15 | -2.566 | -4.385 | -0.525 | -4.385 | 0.789 | -3.109 | 2.549 | -2.566 | -4.385 | -4.385 | -2.566 | -4.385 | -4.385 | -4.385 | -3.109 | -4.385 |
| 16 | -4.385 | -1.437 | 0.713 | -4.385 | -4.385 | -4.385 | -4.385 | -3.109 | -0.703 | 2.5 | -0.842 | -0.92 | -4.385 | -2.566 | -4.385 | -4.385 |
| 17 | -3.109 | -1.197 | -3.109 | -1.958 | 0.185 | 1.434 | -0.022 | 1.813 | -1.096 | 0.158 | -1.197 | -0.374 | -3.109 | -1.582 | -2.216 | -2.216 |
| 18 | -2.216 | -0.64 | -0.09 | -2.566 | 0.846 | 0.648 | 0.185 | -0.842 | -1.752 | 0.041 | -1.752 | -3.109 | -0.64 | 0.237 | 1.597 | -1.096 |
| 19 | -0.92 | 0.01 | 0.262 | -0.842 | -0.842 | 0.01 | 0.443 | 0.56 | -0.77 | 1.132 | -0.126 | 1.022 | -1.958 | -1.197 | -1.197 | -1.958 |
| 20 | -0.842 | -1.31 | -0.581 | -1.752 | 0.503 | -0.09 | 0.483 | 0.211 | 0.31 | 0.262 | -0.126 | -0.703 | -0.328 | 0.101 | 0.859 | -0.09 |