Model info
| Transcription factor | DUX4 (GeneCards) | ||||||||
| Model | DUX4_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 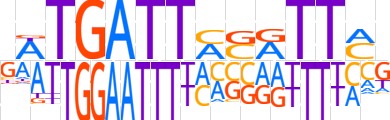 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | KWTGATTASATTCd | ||||||||
| Best auROC (human) | 0.971 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 5 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 367 | ||||||||
| TF family | Paired-related HD factors {3.1.3} | ||||||||
| TF subfamily | DUX {3.1.3.7} | ||||||||
| HGNC | HGNC:50800 | ||||||||
| EntrezGene | GeneID:100288687 (SSTAR profile) | ||||||||
| UniProt ID | DUX4_HUMAN | ||||||||
| UniProt AC | Q9UBX2 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | DUX4 expression | ||||||||
| ReMap ChIP-seq dataset list | DUX4 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 7.0 | 2.0 | 6.0 | 5.0 | 15.0 | 1.0 | 0.0 | 12.0 | 139.0 | 3.0 | 10.0 | 2.0 | 10.0 | 1.0 | 10.0 | 10.0 |
| 02 | 0.0 | 0.0 | 0.0 | 171.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 26.0 | 0.0 | 0.0 | 0.0 | 29.0 |
| 03 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 233.0 | 0.0 |
| 04 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 233.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 0.0 | 233.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 233.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 121.0 | 112.0 | 0.0 | 0.0 |
| 08 | 2.0 | 15.0 | 104.0 | 0.0 | 0.0 | 112.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 2.0 | 0.0 | 127.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 102.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 129.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 104.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 233.0 |
| 12 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 106.0 | 127.0 | 0.0 | 0.0 |
| 13 | 30.0 | 17.0 | 6.0 | 53.0 | 0.0 | 0.0 | 127.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.708 | -1.851 | -0.855 | -1.026 | 0.029 | -2.408 | -3.778 | -0.189 | 2.235 | -1.495 | -0.365 | -1.851 | -0.365 | -2.408 | -0.365 | -0.365 |
| 02 | -3.778 | -3.778 | -3.778 | 2.442 | -3.778 | -3.778 | -3.778 | -0.708 | -3.778 | -3.778 | -3.778 | 0.57 | -3.778 | -3.778 | -3.778 | 0.677 |
| 03 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | 2.751 | -3.778 |
| 04 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | 2.751 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 |
| 05 | -3.778 | -3.778 | -3.778 | 2.751 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 |
| 06 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | 2.751 |
| 07 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | 2.097 | 2.02 | -3.778 | -3.778 |
| 08 | -1.851 | 0.029 | 1.946 | -3.778 | -3.778 | 2.02 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 |
| 09 | -3.778 | -3.778 | -1.851 | -3.778 | 2.145 | -3.778 | -3.778 | -3.778 | -1.851 | -3.778 | 1.927 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 |
| 10 | -3.778 | -3.778 | -3.778 | 2.161 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | 1.946 | -3.778 | -3.778 | -3.778 | -3.778 |
| 11 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | 2.751 |
| 12 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | 1.965 | 2.145 | -3.778 | -3.778 |
| 13 | 0.711 | 0.151 | -0.855 | 1.275 | -3.778 | -3.778 | 2.145 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 | -3.778 |