Model info
| Transcription factor | E2F4 (GeneCards) | ||||||||
| Model | E2F4_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 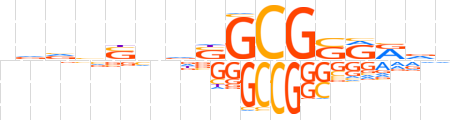 | ||||||||
| LOGO (reverse complement) | 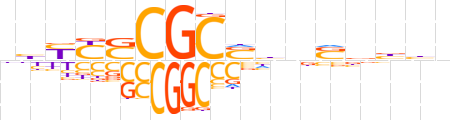 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndvvSndSGCGSRRdn | ||||||||
| Best auROC (human) | 0.963 | ||||||||
| Best auROC (mouse) | 0.989 | ||||||||
| Peak sets in benchmark (human) | 15 | ||||||||
| Peak sets in benchmark (mouse) | 37 | ||||||||
| Aligned words | 458 | ||||||||
| TF family | E2F-related factors {3.3.2} | ||||||||
| TF subfamily | E2F {3.3.2.1} | ||||||||
| HGNC | HGNC:3118 | ||||||||
| EntrezGene | GeneID:1874 (SSTAR profile) | ||||||||
| UniProt ID | E2F4_HUMAN | ||||||||
| UniProt AC | Q16254 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | E2F4 expression | ||||||||
| ReMap ChIP-seq dataset list | E2F4 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 40.0 | 13.0 | 31.0 | 10.0 | 31.0 | 17.0 | 43.0 | 27.0 | 33.0 | 21.0 | 78.0 | 35.0 | 12.0 | 7.0 | 39.0 | 16.0 |
| 02 | 55.0 | 18.0 | 39.0 | 4.0 | 22.0 | 11.0 | 18.0 | 7.0 | 70.0 | 29.0 | 75.0 | 17.0 | 17.0 | 15.0 | 48.0 | 8.0 |
| 03 | 55.0 | 47.0 | 50.0 | 12.0 | 11.0 | 20.0 | 21.0 | 21.0 | 33.0 | 92.0 | 38.0 | 17.0 | 6.0 | 12.0 | 17.0 | 1.0 |
| 04 | 8.0 | 14.0 | 67.0 | 16.0 | 4.0 | 36.0 | 110.0 | 21.0 | 8.0 | 58.0 | 54.0 | 6.0 | 2.0 | 11.0 | 27.0 | 11.0 |
| 05 | 10.0 | 4.0 | 7.0 | 1.0 | 38.0 | 18.0 | 39.0 | 24.0 | 51.0 | 133.0 | 46.0 | 28.0 | 19.0 | 6.0 | 14.0 | 15.0 |
| 06 | 25.0 | 12.0 | 50.0 | 31.0 | 62.0 | 18.0 | 61.0 | 20.0 | 31.0 | 16.0 | 44.0 | 15.0 | 15.0 | 10.0 | 20.0 | 23.0 |
| 07 | 9.0 | 13.0 | 81.0 | 30.0 | 11.0 | 5.0 | 35.0 | 5.0 | 12.0 | 63.0 | 84.0 | 16.0 | 1.0 | 7.0 | 78.0 | 3.0 |
| 08 | 3.0 | 2.0 | 28.0 | 0.0 | 3.0 | 5.0 | 80.0 | 0.0 | 5.0 | 15.0 | 258.0 | 0.0 | 0.0 | 4.0 | 50.0 | 0.0 |
| 09 | 0.0 | 11.0 | 0.0 | 0.0 | 0.0 | 25.0 | 0.0 | 1.0 | 2.0 | 407.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 2.0 | 0.0 | 2.0 | 4.0 | 432.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 7.0 | 0.0 |
| 11 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 3.0 | 1.0 | 1.0 | 12.0 | 186.0 | 234.0 | 9.0 | 0.0 | 0.0 | 5.0 | 0.0 |
| 12 | 2.0 | 0.0 | 11.0 | 0.0 | 43.0 | 32.0 | 110.0 | 4.0 | 29.0 | 22.0 | 185.0 | 5.0 | 4.0 | 1.0 | 4.0 | 1.0 |
| 13 | 36.0 | 4.0 | 35.0 | 3.0 | 40.0 | 1.0 | 12.0 | 2.0 | 189.0 | 30.0 | 78.0 | 13.0 | 3.0 | 1.0 | 5.0 | 1.0 |
| 14 | 139.0 | 22.0 | 92.0 | 15.0 | 14.0 | 6.0 | 8.0 | 8.0 | 45.0 | 22.0 | 41.0 | 22.0 | 2.0 | 2.0 | 8.0 | 7.0 |
| 15 | 79.0 | 37.0 | 63.0 | 21.0 | 6.0 | 18.0 | 16.0 | 12.0 | 45.0 | 54.0 | 31.0 | 19.0 | 3.0 | 3.0 | 28.0 | 18.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.342 | -0.763 | 0.09 | -1.017 | 0.09 | -0.501 | 0.413 | -0.047 | 0.151 | -0.294 | 1.005 | 0.209 | -0.84 | -1.358 | 0.317 | -0.561 |
| 02 | 0.658 | -0.445 | 0.317 | -1.879 | -0.248 | -0.925 | -0.445 | -1.358 | 0.897 | 0.024 | 0.966 | -0.501 | -0.501 | -0.624 | 0.522 | -1.231 |
| 03 | 0.658 | 0.502 | 0.563 | -0.84 | -0.925 | -0.342 | -0.294 | -0.294 | 0.151 | 1.169 | 0.291 | -0.501 | -1.503 | -0.84 | -0.501 | -3.033 |
| 04 | -1.231 | -0.691 | 0.854 | -0.561 | -1.879 | 0.237 | 1.347 | -0.294 | -1.231 | 0.71 | 0.639 | -1.503 | -2.489 | -0.925 | -0.047 | -0.925 |
| 05 | -1.017 | -1.879 | -1.358 | -3.033 | 0.291 | -0.445 | 0.317 | -0.163 | 0.583 | 1.537 | 0.48 | -0.011 | -0.392 | -1.503 | -0.691 | -0.624 |
| 06 | -0.123 | -0.84 | 0.563 | 0.09 | 0.777 | -0.445 | 0.76 | -0.342 | 0.09 | -0.561 | 0.436 | -0.624 | -0.624 | -1.017 | -0.342 | -0.205 |
| 07 | -1.118 | -0.763 | 1.042 | 0.057 | -0.925 | -1.674 | 0.209 | -1.674 | -0.84 | 0.792 | 1.079 | -0.561 | -3.033 | -1.358 | 1.005 | -2.138 |
| 08 | -2.138 | -2.489 | -0.011 | -4.318 | -2.138 | -1.674 | 1.03 | -4.318 | -1.674 | -0.624 | 2.198 | -4.318 | -4.318 | -1.879 | 0.563 | -4.318 |
| 09 | -4.318 | -0.925 | -4.318 | -4.318 | -4.318 | -0.123 | -4.318 | -3.033 | -2.489 | 2.653 | -4.318 | -1.358 | -4.318 | -4.318 | -4.318 | -4.318 |
| 10 | -4.318 | -4.318 | -2.489 | -4.318 | -2.489 | -1.879 | 2.713 | -1.674 | -4.318 | -4.318 | -4.318 | -4.318 | -4.318 | -3.033 | -1.358 | -4.318 |
| 11 | -3.033 | -4.318 | -3.033 | -4.318 | -4.318 | -2.138 | -3.033 | -3.033 | -0.84 | 1.871 | 2.1 | -1.118 | -4.318 | -4.318 | -1.674 | -4.318 |
| 12 | -2.489 | -4.318 | -0.925 | -4.318 | 0.413 | 0.121 | 1.347 | -1.879 | 0.024 | -0.248 | 1.866 | -1.674 | -1.879 | -3.033 | -1.879 | -3.033 |
| 13 | 0.237 | -1.879 | 0.209 | -2.138 | 0.342 | -3.033 | -0.84 | -2.489 | 1.887 | 0.057 | 1.005 | -0.763 | -2.138 | -3.033 | -1.674 | -3.033 |
| 14 | 1.581 | -0.248 | 1.169 | -0.624 | -0.691 | -1.503 | -1.231 | -1.231 | 0.458 | -0.248 | 0.366 | -0.248 | -2.489 | -2.489 | -1.231 | -1.358 |
| 15 | 1.018 | 0.264 | 0.792 | -0.294 | -1.503 | -0.445 | -0.561 | -0.84 | 0.458 | 0.639 | 0.09 | -0.392 | -2.138 | -2.138 | -0.011 | -0.445 |