Model info
| Transcription factor | E2f4 | ||||||||
| Model | E2F4_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 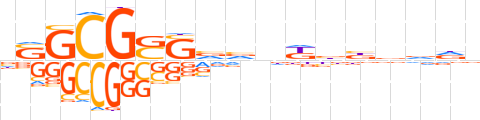 | ||||||||
| LOGO (reverse complement) | 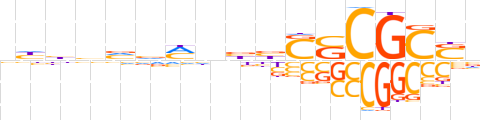 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nSGCGSSvvnKbbvvdn | ||||||||
| Best auROC (human) | 0.957 | ||||||||
| Best auROC (mouse) | 0.989 | ||||||||
| Peak sets in benchmark (human) | 15 | ||||||||
| Peak sets in benchmark (mouse) | 37 | ||||||||
| Aligned words | 487 | ||||||||
| TF family | E2F-related factors {3.3.2} | ||||||||
| TF subfamily | E2F {3.3.2.1} | ||||||||
| MGI | MGI:103012 | ||||||||
| EntrezGene | GeneID:104394 (SSTAR profile) | ||||||||
| UniProt ID | E2F4_MOUSE | ||||||||
| UniProt AC | Q8R0K9 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | E2f4 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 13.0 | 8.0 | 63.0 | 3.0 | 23.0 | 23.0 | 87.0 | 10.0 | 14.0 | 56.0 | 103.0 | 1.0 | 5.0 | 17.0 | 57.0 | 3.0 |
| 02 | 1.0 | 6.0 | 48.0 | 0.0 | 0.0 | 24.0 | 77.0 | 3.0 | 3.0 | 39.0 | 262.0 | 6.0 | 0.0 | 2.0 | 13.0 | 2.0 |
| 03 | 1.0 | 3.0 | 0.0 | 0.0 | 6.0 | 65.0 | 0.0 | 0.0 | 25.0 | 371.0 | 0.0 | 4.0 | 1.0 | 10.0 | 0.0 | 0.0 |
| 04 | 0.0 | 0.0 | 32.0 | 1.0 | 3.0 | 1.0 | 435.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 1.0 |
| 05 | 1.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 4.0 | 234.0 | 221.0 | 10.0 | 1.0 | 5.0 | 4.0 | 2.0 |
| 06 | 0.0 | 1.0 | 5.0 | 0.0 | 9.0 | 46.0 | 166.0 | 18.0 | 13.0 | 24.0 | 187.0 | 5.0 | 0.0 | 2.0 | 10.0 | 0.0 |
| 07 | 3.0 | 8.0 | 8.0 | 3.0 | 36.0 | 12.0 | 23.0 | 2.0 | 136.0 | 95.0 | 130.0 | 7.0 | 5.0 | 4.0 | 13.0 | 1.0 |
| 08 | 99.0 | 19.0 | 57.0 | 5.0 | 45.0 | 23.0 | 37.0 | 14.0 | 70.0 | 59.0 | 31.0 | 14.0 | 8.0 | 4.0 | 1.0 | 0.0 |
| 09 | 78.0 | 40.0 | 71.0 | 33.0 | 15.0 | 19.0 | 30.0 | 41.0 | 48.0 | 25.0 | 30.0 | 23.0 | 1.0 | 11.0 | 9.0 | 12.0 |
| 10 | 35.0 | 3.0 | 58.0 | 46.0 | 11.0 | 3.0 | 41.0 | 40.0 | 9.0 | 3.0 | 80.0 | 48.0 | 1.0 | 5.0 | 32.0 | 71.0 |
| 11 | 7.0 | 10.0 | 31.0 | 8.0 | 3.0 | 5.0 | 1.0 | 5.0 | 20.0 | 89.0 | 60.0 | 42.0 | 13.0 | 36.0 | 44.0 | 112.0 |
| 12 | 6.0 | 3.0 | 29.0 | 5.0 | 5.0 | 28.0 | 73.0 | 34.0 | 10.0 | 28.0 | 83.0 | 15.0 | 2.0 | 50.0 | 41.0 | 74.0 |
| 13 | 11.0 | 8.0 | 3.0 | 1.0 | 24.0 | 18.0 | 52.0 | 15.0 | 40.0 | 72.0 | 80.0 | 34.0 | 23.0 | 29.0 | 60.0 | 16.0 |
| 14 | 63.0 | 5.0 | 25.0 | 5.0 | 37.0 | 14.0 | 59.0 | 17.0 | 84.0 | 49.0 | 43.0 | 19.0 | 2.0 | 15.0 | 34.0 | 15.0 |
| 15 | 92.0 | 8.0 | 66.0 | 20.0 | 14.0 | 4.0 | 47.0 | 18.0 | 54.0 | 11.0 | 77.0 | 19.0 | 10.0 | 7.0 | 18.0 | 21.0 |
| 16 | 53.0 | 43.0 | 54.0 | 20.0 | 4.0 | 3.0 | 16.0 | 7.0 | 32.0 | 64.0 | 82.0 | 30.0 | 5.0 | 24.0 | 21.0 | 28.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.832 | -1.3 | 0.723 | -2.206 | -0.274 | -0.274 | 1.044 | -1.086 | -0.76 | 0.606 | 1.212 | -3.099 | -1.742 | -0.571 | 0.624 | -2.206 |
| 02 | -3.099 | -1.572 | 0.453 | -4.377 | -4.377 | -0.232 | 0.923 | -2.206 | -2.206 | 0.247 | 2.144 | -1.572 | -4.377 | -2.556 | -0.832 | -2.556 |
| 03 | -3.099 | -2.206 | -4.377 | -4.377 | -1.572 | 0.754 | -4.377 | -4.377 | -0.192 | 2.491 | -4.377 | -1.948 | -3.099 | -1.086 | -4.377 | -4.377 |
| 04 | -4.377 | -4.377 | 0.051 | -3.099 | -2.206 | -3.099 | 2.65 | -1.086 | -4.377 | -4.377 | -4.377 | -4.377 | -3.099 | -4.377 | -2.556 | -3.099 |
| 05 | -3.099 | -4.377 | -2.206 | -4.377 | -4.377 | -4.377 | -3.099 | -4.377 | -1.948 | 2.031 | 1.974 | -1.086 | -3.099 | -1.742 | -1.948 | -2.556 |
| 06 | -4.377 | -3.099 | -1.742 | -4.377 | -1.187 | 0.411 | 1.688 | -0.515 | -0.832 | -0.232 | 1.807 | -1.742 | -4.377 | -2.556 | -1.086 | -4.377 |
| 07 | -2.206 | -1.3 | -1.3 | -2.206 | 0.168 | -0.91 | -0.274 | -2.556 | 1.489 | 1.132 | 1.444 | -1.427 | -1.742 | -1.948 | -0.832 | -3.099 |
| 08 | 1.173 | -0.462 | 0.624 | -1.742 | 0.389 | -0.274 | 0.195 | -0.76 | 0.828 | 0.658 | 0.02 | -0.76 | -1.3 | -1.948 | -3.099 | -4.377 |
| 09 | 0.935 | 0.272 | 0.842 | 0.082 | -0.693 | -0.462 | -0.012 | 0.297 | 0.453 | -0.192 | -0.012 | -0.274 | -3.099 | -0.994 | -1.187 | -0.91 |
| 10 | 0.14 | -2.206 | 0.641 | 0.411 | -0.994 | -2.206 | 0.297 | 0.272 | -1.187 | -2.206 | 0.961 | 0.453 | -3.099 | -1.742 | 0.051 | 0.842 |
| 11 | -1.427 | -1.086 | 0.02 | -1.3 | -2.206 | -1.742 | -3.099 | -1.742 | -0.411 | 1.067 | 0.674 | 0.321 | -0.832 | 0.168 | 0.367 | 1.296 |
| 12 | -1.572 | -2.206 | -0.046 | -1.742 | -1.742 | -0.08 | 0.869 | 0.111 | -1.086 | -0.08 | 0.997 | -0.693 | -2.556 | 0.493 | 0.297 | 0.883 |
| 13 | -0.994 | -1.3 | -2.206 | -3.099 | -0.232 | -0.515 | 0.532 | -0.693 | 0.272 | 0.856 | 0.961 | 0.111 | -0.274 | -0.046 | 0.674 | -0.63 |
| 14 | 0.723 | -1.742 | -0.192 | -1.742 | 0.195 | -0.76 | 0.658 | -0.571 | 1.009 | 0.473 | 0.344 | -0.462 | -2.556 | -0.693 | 0.111 | -0.693 |
| 15 | 1.1 | -1.3 | 0.769 | -0.411 | -0.76 | -1.948 | 0.432 | -0.515 | 0.57 | -0.994 | 0.923 | -0.462 | -1.086 | -1.427 | -0.515 | -0.364 |
| 16 | 0.551 | 0.344 | 0.57 | -0.411 | -1.948 | -2.206 | -0.63 | -1.427 | 0.051 | 0.739 | 0.985 | -0.012 | -1.742 | -0.232 | -0.364 | -0.08 |