Model info
| Transcription factor | EGR1 (GeneCards) | ||||||||
| Model | EGR1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 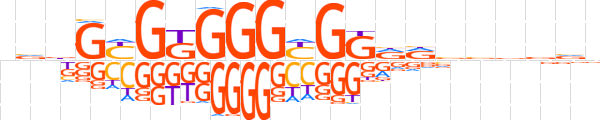 | ||||||||
| LOGO (reverse complement) | 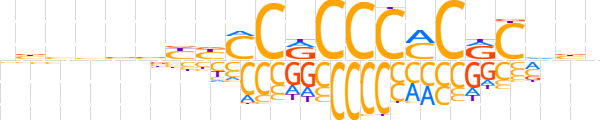 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvbGYGKGGGYGGRRvbvbvv | ||||||||
| Best auROC (human) | 0.991 | ||||||||
| Best auROC (mouse) | 0.986 | ||||||||
| Peak sets in benchmark (human) | 36 | ||||||||
| Peak sets in benchmark (mouse) | 12 | ||||||||
| Aligned words | 526 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | EGR factors {2.3.1.3} | ||||||||
| HGNC | HGNC:3238 | ||||||||
| EntrezGene | GeneID:1958 (SSTAR profile) | ||||||||
| UniProt ID | EGR1_HUMAN | ||||||||
| UniProt AC | P18146 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | EGR1 expression | ||||||||
| ReMap ChIP-seq dataset list | EGR1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 22.0 | 17.0 | 50.0 | 8.0 | 26.0 | 20.0 | 41.0 | 14.0 | 60.0 | 35.0 | 85.0 | 14.0 | 5.0 | 19.0 | 59.0 | 15.0 |
| 02 | 15.0 | 26.0 | 48.0 | 24.0 | 9.0 | 18.0 | 20.0 | 44.0 | 27.0 | 44.0 | 79.0 | 85.0 | 9.0 | 8.0 | 16.0 | 18.0 |
| 03 | 2.0 | 4.0 | 54.0 | 0.0 | 6.0 | 6.0 | 79.0 | 5.0 | 10.0 | 3.0 | 148.0 | 2.0 | 7.0 | 6.0 | 154.0 | 4.0 |
| 04 | 4.0 | 12.0 | 2.0 | 7.0 | 3.0 | 10.0 | 0.0 | 6.0 | 65.0 | 294.0 | 3.0 | 73.0 | 2.0 | 6.0 | 0.0 | 3.0 |
| 05 | 1.0 | 0.0 | 73.0 | 0.0 | 1.0 | 0.0 | 321.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 89.0 | 0.0 |
| 06 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 270.0 | 216.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 19.0 | 1.0 | 251.0 | 0.0 | 0.0 | 0.0 | 217.0 | 0.0 |
| 08 | 0.0 | 0.0 | 19.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 466.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 486.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 72.0 | 302.0 | 0.0 | 116.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 1.0 | 0.0 | 71.0 | 0.0 | 0.0 | 4.0 | 297.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 115.0 | 0.0 |
| 12 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 2.0 | 0.0 | 12.0 | 14.0 | 377.0 | 80.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 13 | 2.0 | 2.0 | 9.0 | 0.0 | 1.0 | 4.0 | 7.0 | 4.0 | 121.0 | 29.0 | 201.0 | 30.0 | 3.0 | 8.0 | 60.0 | 9.0 |
| 14 | 23.0 | 9.0 | 87.0 | 8.0 | 7.0 | 5.0 | 19.0 | 12.0 | 63.0 | 21.0 | 165.0 | 28.0 | 10.0 | 4.0 | 26.0 | 3.0 |
| 15 | 15.0 | 32.0 | 51.0 | 5.0 | 15.0 | 11.0 | 8.0 | 5.0 | 59.0 | 106.0 | 94.0 | 38.0 | 6.0 | 14.0 | 26.0 | 5.0 |
| 16 | 13.0 | 23.0 | 47.0 | 12.0 | 14.0 | 61.0 | 54.0 | 34.0 | 36.0 | 48.0 | 65.0 | 30.0 | 4.0 | 19.0 | 20.0 | 10.0 |
| 17 | 13.0 | 10.0 | 37.0 | 7.0 | 32.0 | 37.0 | 47.0 | 35.0 | 41.0 | 42.0 | 70.0 | 33.0 | 11.0 | 11.0 | 46.0 | 18.0 |
| 18 | 22.0 | 14.0 | 41.0 | 20.0 | 20.0 | 26.0 | 37.0 | 17.0 | 35.0 | 30.0 | 97.0 | 38.0 | 6.0 | 20.0 | 56.0 | 11.0 |
| 19 | 15.0 | 13.0 | 49.0 | 6.0 | 15.0 | 26.0 | 30.0 | 19.0 | 60.0 | 33.0 | 113.0 | 25.0 | 4.0 | 15.0 | 50.0 | 17.0 |
| 20 | 25.0 | 10.0 | 50.0 | 9.0 | 21.0 | 17.0 | 29.0 | 20.0 | 48.0 | 53.0 | 108.0 | 33.0 | 4.0 | 16.0 | 28.0 | 19.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.326 | -0.579 | 0.485 | -1.308 | -0.162 | -0.419 | 0.289 | -0.768 | 0.666 | 0.132 | 1.013 | -0.768 | -1.75 | -0.47 | 0.65 | -0.701 |
| 02 | -0.701 | -0.162 | 0.445 | -0.24 | -1.195 | -0.523 | -0.419 | 0.359 | -0.124 | 0.359 | 0.94 | 1.013 | -1.195 | -1.308 | -0.638 | -0.523 |
| 03 | -2.564 | -1.956 | 0.562 | -4.383 | -1.58 | -1.58 | 0.94 | -1.75 | -1.094 | -2.214 | 1.565 | -2.564 | -1.435 | -1.58 | 1.605 | -1.956 |
| 04 | -1.956 | -0.918 | -2.564 | -1.435 | -2.214 | -1.094 | -4.383 | -1.58 | 0.746 | 2.251 | -2.214 | 0.861 | -2.564 | -1.58 | -4.383 | -2.214 |
| 05 | -3.107 | -4.383 | 0.861 | -4.383 | -3.107 | -4.383 | 2.338 | -4.383 | -4.383 | -4.383 | -1.75 | -4.383 | -4.383 | -4.383 | 1.059 | -4.383 |
| 06 | -4.383 | -4.383 | -3.107 | -3.107 | -4.383 | -4.383 | -4.383 | -4.383 | -3.107 | -3.107 | 2.165 | 1.943 | -4.383 | -4.383 | -4.383 | -4.383 |
| 07 | -4.383 | -4.383 | -3.107 | -4.383 | -4.383 | -4.383 | -3.107 | -4.383 | -0.47 | -3.107 | 2.093 | -4.383 | -4.383 | -4.383 | 1.947 | -4.383 |
| 08 | -4.383 | -4.383 | -0.47 | -4.383 | -4.383 | -4.383 | -3.107 | -4.383 | -4.383 | -3.107 | 2.711 | -2.214 | -4.383 | -4.383 | -4.383 | -4.383 |
| 09 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | -3.107 | -4.383 | -4.383 | -4.383 | 2.753 | -4.383 | -4.383 | -4.383 | -2.214 | -4.383 |
| 10 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | 0.848 | 2.277 | -4.383 | 1.323 | -4.383 | -4.383 | -4.383 | -4.383 |
| 11 | -3.107 | -4.383 | 0.834 | -4.383 | -4.383 | -1.956 | 2.261 | -3.107 | -4.383 | -4.383 | -4.383 | -4.383 | -3.107 | -4.383 | 1.314 | -4.383 |
| 12 | -4.383 | -3.107 | -3.107 | -4.383 | -3.107 | -3.107 | -2.564 | -4.383 | -0.918 | -0.768 | 2.499 | 0.952 | -4.383 | -4.383 | -3.107 | -4.383 |
| 13 | -2.564 | -2.564 | -1.195 | -4.383 | -3.107 | -1.956 | -1.435 | -1.956 | 1.365 | -0.054 | 1.871 | -0.02 | -2.214 | -1.308 | 0.666 | -1.195 |
| 14 | -0.282 | -1.195 | 1.036 | -1.308 | -1.435 | -1.75 | -0.47 | -0.918 | 0.715 | -0.372 | 1.674 | -0.088 | -1.094 | -1.956 | -0.162 | -2.214 |
| 15 | -0.701 | 0.043 | 0.505 | -1.75 | -0.701 | -1.002 | -1.308 | -1.75 | 0.65 | 1.233 | 1.113 | 0.213 | -1.58 | -0.768 | -0.162 | -1.75 |
| 16 | -0.84 | -0.282 | 0.424 | -0.918 | -0.768 | 0.683 | 0.562 | 0.103 | 0.16 | 0.445 | 0.746 | -0.02 | -1.956 | -0.47 | -0.419 | -1.094 |
| 17 | -0.84 | -1.094 | 0.187 | -1.435 | 0.043 | 0.187 | 0.424 | 0.132 | 0.289 | 0.312 | 0.82 | 0.074 | -1.002 | -1.002 | 0.403 | -0.523 |
| 18 | -0.326 | -0.768 | 0.289 | -0.419 | -0.419 | -0.162 | 0.187 | -0.579 | 0.132 | -0.02 | 1.144 | 0.213 | -1.58 | -0.419 | 0.598 | -1.002 |
| 19 | -0.701 | -0.84 | 0.465 | -1.58 | -0.701 | -0.162 | -0.02 | -0.47 | 0.666 | 0.074 | 1.296 | -0.2 | -1.956 | -0.701 | 0.485 | -0.579 |
| 20 | -0.2 | -1.094 | 0.485 | -1.195 | -0.372 | -0.579 | -0.054 | -0.419 | 0.445 | 0.543 | 1.251 | 0.074 | -1.956 | -0.638 | -0.088 | -0.47 |