Model info
| Transcription factor | EGR2 (GeneCards) | ||||||||
| Model | EGR2_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 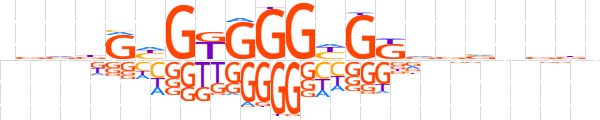 | ||||||||
| LOGO (reverse complement) | 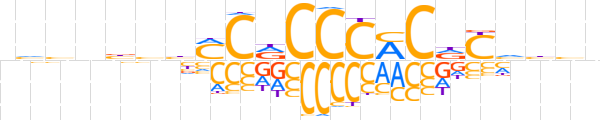 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vdvdGhGKGGGYGKvdvnddn | ||||||||
| Best auROC (human) | 0.989 | ||||||||
| Best auROC (mouse) | 0.987 | ||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | 25 | ||||||||
| Aligned words | 625 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | EGR factors {2.3.1.3} | ||||||||
| HGNC | HGNC:3239 | ||||||||
| EntrezGene | GeneID:1959 (SSTAR profile) | ||||||||
| UniProt ID | EGR2_HUMAN | ||||||||
| UniProt AC | P11161 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | EGR2 expression | ||||||||
| ReMap ChIP-seq dataset list | EGR2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 25.0 | 17.0 | 37.0 | 11.0 | 25.0 | 21.0 | 33.0 | 20.0 | 42.0 | 24.0 | 102.0 | 45.0 | 11.0 | 13.0 | 33.0 | 14.0 |
| 02 | 35.0 | 23.0 | 32.0 | 13.0 | 17.0 | 21.0 | 21.0 | 16.0 | 66.0 | 41.0 | 83.0 | 15.0 | 12.0 | 13.0 | 60.0 | 5.0 |
| 03 | 20.0 | 13.0 | 70.0 | 27.0 | 10.0 | 18.0 | 28.0 | 42.0 | 28.0 | 23.0 | 71.0 | 74.0 | 12.0 | 7.0 | 19.0 | 11.0 |
| 04 | 12.0 | 4.0 | 53.0 | 1.0 | 13.0 | 10.0 | 36.0 | 2.0 | 17.0 | 8.0 | 158.0 | 5.0 | 9.0 | 11.0 | 131.0 | 3.0 |
| 05 | 18.0 | 18.0 | 3.0 | 12.0 | 5.0 | 10.0 | 1.0 | 17.0 | 87.0 | 191.0 | 3.0 | 97.0 | 2.0 | 5.0 | 1.0 | 3.0 |
| 06 | 1.0 | 1.0 | 109.0 | 1.0 | 0.0 | 1.0 | 221.0 | 2.0 | 0.0 | 1.0 | 7.0 | 0.0 | 2.0 | 0.0 | 126.0 | 1.0 |
| 07 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 0.0 | 3.0 | 4.0 | 4.0 | 165.0 | 290.0 | 1.0 | 0.0 | 1.0 | 2.0 |
| 08 | 0.0 | 0.0 | 4.0 | 1.0 | 0.0 | 0.0 | 4.0 | 0.0 | 33.0 | 1.0 | 130.0 | 3.0 | 11.0 | 2.0 | 281.0 | 3.0 |
| 09 | 0.0 | 0.0 | 44.0 | 0.0 | 0.0 | 0.0 | 2.0 | 1.0 | 1.0 | 3.0 | 402.0 | 13.0 | 0.0 | 0.0 | 7.0 | 0.0 |
| 10 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 3.0 | 449.0 | 3.0 | 0.0 | 0.0 | 14.0 | 0.0 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 1.0 | 0.0 | 1.0 | 91.0 | 229.0 | 0.0 | 146.0 | 0.0 | 2.0 | 1.0 | 0.0 |
| 12 | 3.0 | 0.0 | 88.0 | 2.0 | 9.0 | 0.0 | 215.0 | 8.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 | 136.0 | 9.0 |
| 13 | 1.0 | 1.0 | 10.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 26.0 | 14.0 | 299.0 | 101.0 | 1.0 | 0.0 | 13.0 | 5.0 |
| 14 | 10.0 | 4.0 | 13.0 | 1.0 | 0.0 | 2.0 | 8.0 | 5.0 | 105.0 | 55.0 | 122.0 | 40.0 | 18.0 | 15.0 | 61.0 | 14.0 |
| 15 | 27.0 | 8.0 | 81.0 | 17.0 | 24.0 | 17.0 | 14.0 | 21.0 | 44.0 | 24.0 | 74.0 | 62.0 | 10.0 | 11.0 | 30.0 | 9.0 |
| 16 | 25.0 | 13.0 | 55.0 | 12.0 | 16.0 | 14.0 | 17.0 | 13.0 | 40.0 | 42.0 | 93.0 | 24.0 | 10.0 | 17.0 | 70.0 | 12.0 |
| 17 | 25.0 | 14.0 | 38.0 | 14.0 | 18.0 | 18.0 | 21.0 | 29.0 | 59.0 | 42.0 | 67.0 | 67.0 | 3.0 | 13.0 | 34.0 | 11.0 |
| 18 | 23.0 | 11.0 | 52.0 | 19.0 | 22.0 | 22.0 | 22.0 | 21.0 | 30.0 | 31.0 | 64.0 | 35.0 | 13.0 | 15.0 | 81.0 | 12.0 |
| 19 | 21.0 | 13.0 | 38.0 | 16.0 | 22.0 | 10.0 | 22.0 | 25.0 | 25.0 | 32.0 | 104.0 | 58.0 | 20.0 | 6.0 | 49.0 | 12.0 |
| 20 | 12.0 | 29.0 | 38.0 | 9.0 | 16.0 | 12.0 | 20.0 | 13.0 | 61.0 | 51.0 | 67.0 | 34.0 | 12.0 | 18.0 | 57.0 | 24.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.165 | -0.544 | 0.222 | -0.967 | -0.165 | -0.337 | 0.109 | -0.385 | 0.347 | -0.205 | 1.229 | 0.416 | -0.967 | -0.805 | 0.109 | -0.733 |
| 02 | 0.167 | -0.247 | 0.078 | -0.805 | -0.544 | -0.337 | -0.337 | -0.603 | 0.796 | 0.323 | 1.024 | -0.666 | -0.883 | -0.805 | 0.701 | -1.716 |
| 03 | -0.385 | -0.805 | 0.855 | -0.089 | -1.059 | -0.488 | -0.054 | 0.347 | -0.054 | -0.247 | 0.869 | 0.91 | -0.883 | -1.4 | -0.435 | -0.967 |
| 04 | -0.883 | -1.921 | 0.578 | -3.074 | -0.805 | -1.059 | 0.195 | -2.53 | -0.544 | -1.273 | 1.666 | -1.716 | -1.16 | -0.967 | 1.479 | -2.18 |
| 05 | -0.488 | -0.488 | -2.18 | -0.883 | -1.716 | -1.059 | -3.074 | -0.544 | 1.071 | 1.855 | -2.18 | 1.179 | -2.53 | -1.716 | -3.074 | -2.18 |
| 06 | -3.074 | -3.074 | 1.295 | -3.074 | -4.354 | -3.074 | 2.0 | -2.53 | -4.354 | -3.074 | -1.4 | -4.354 | -2.53 | -4.354 | 1.44 | -3.074 |
| 07 | -4.354 | -4.354 | -3.074 | -2.53 | -4.354 | -4.354 | -4.354 | -2.18 | -1.921 | -1.921 | 1.709 | 2.272 | -3.074 | -4.354 | -3.074 | -2.53 |
| 08 | -4.354 | -4.354 | -1.921 | -3.074 | -4.354 | -4.354 | -1.921 | -4.354 | 0.109 | -3.074 | 1.471 | -2.18 | -0.967 | -2.53 | 2.24 | -2.18 |
| 09 | -4.354 | -4.354 | 0.393 | -4.354 | -4.354 | -4.354 | -2.53 | -3.074 | -3.074 | -2.18 | 2.598 | -0.805 | -4.354 | -4.354 | -1.4 | -4.354 |
| 10 | -4.354 | -4.354 | -3.074 | -4.354 | -4.354 | -3.074 | -2.53 | -4.354 | -4.354 | -2.18 | 2.708 | -2.18 | -4.354 | -4.354 | -0.733 | -4.354 |
| 11 | -4.354 | -4.354 | -4.354 | -4.354 | -2.53 | -3.074 | -4.354 | -3.074 | 1.116 | 2.036 | -4.354 | 1.587 | -4.354 | -2.53 | -3.074 | -4.354 |
| 12 | -2.18 | -4.354 | 1.082 | -2.53 | -1.16 | -4.354 | 1.973 | -1.273 | -4.354 | -4.354 | -3.074 | -4.354 | -2.53 | -4.354 | 1.516 | -1.16 |
| 13 | -3.074 | -3.074 | -1.059 | -2.53 | -4.354 | -4.354 | -4.354 | -4.354 | -0.127 | -0.733 | 2.302 | 1.219 | -3.074 | -4.354 | -0.805 | -1.716 |
| 14 | -1.059 | -1.921 | -0.805 | -3.074 | -4.354 | -2.53 | -1.273 | -1.716 | 1.258 | 0.615 | 1.408 | 0.299 | -0.488 | -0.666 | 0.718 | -0.733 |
| 15 | -0.089 | -1.273 | 1.0 | -0.544 | -0.205 | -0.544 | -0.733 | -0.337 | 0.393 | -0.205 | 0.91 | 0.734 | -1.059 | -0.967 | 0.015 | -1.16 |
| 16 | -0.165 | -0.805 | 0.615 | -0.883 | -0.603 | -0.733 | -0.544 | -0.805 | 0.299 | 0.347 | 1.137 | -0.205 | -1.059 | -0.544 | 0.855 | -0.883 |
| 17 | -0.165 | -0.733 | 0.248 | -0.733 | -0.488 | -0.488 | -0.337 | -0.019 | 0.685 | 0.347 | 0.811 | 0.811 | -2.18 | -0.805 | 0.138 | -0.967 |
| 18 | -0.247 | -0.967 | 0.559 | -0.435 | -0.291 | -0.291 | -0.291 | -0.337 | 0.015 | 0.047 | 0.765 | 0.167 | -0.805 | -0.666 | 1.0 | -0.883 |
| 19 | -0.337 | -0.805 | 0.248 | -0.603 | -0.291 | -1.059 | -0.291 | -0.165 | -0.165 | 0.078 | 1.249 | 0.668 | -0.385 | -1.546 | 0.5 | -0.883 |
| 20 | -0.883 | -0.019 | 0.248 | -1.16 | -0.603 | -0.883 | -0.385 | -0.805 | 0.718 | 0.54 | 0.811 | 0.138 | -0.883 | -0.488 | 0.65 | -0.205 |