Model info
| Transcription factor | ELF1 (GeneCards) | ||||||||
| Model | ELF1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 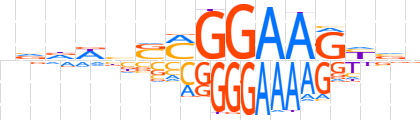 | ||||||||
| LOGO (reverse complement) | 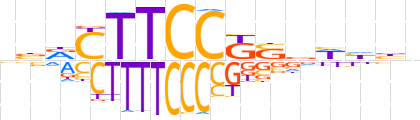 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvdWbSMGGAAGYvv | ||||||||
| Best auROC (human) | 0.992 | ||||||||
| Best auROC (mouse) | 0.977 | ||||||||
| Peak sets in benchmark (human) | 43 | ||||||||
| Peak sets in benchmark (mouse) | 5 | ||||||||
| Aligned words | 403 | ||||||||
| TF family | Ets-related factors {3.5.2} | ||||||||
| TF subfamily | Elf-1-like factors {3.5.2.3} | ||||||||
| HGNC | HGNC:3316 | ||||||||
| EntrezGene | GeneID:1997 (SSTAR profile) | ||||||||
| UniProt ID | ELF1_HUMAN | ||||||||
| UniProt AC | P32519 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ELF1 expression | ||||||||
| ReMap ChIP-seq dataset list | ELF1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 28.0 | 19.0 | 42.0 | 7.0 | 38.0 | 17.0 | 42.0 | 15.0 | 26.0 | 20.0 | 80.0 | 13.0 | 7.0 | 10.0 | 27.0 | 12.0 |
| 02 | 46.0 | 15.0 | 22.0 | 16.0 | 30.0 | 13.0 | 12.0 | 11.0 | 115.0 | 19.0 | 36.0 | 21.0 | 16.0 | 7.0 | 15.0 | 9.0 |
| 03 | 148.0 | 14.0 | 26.0 | 19.0 | 24.0 | 2.0 | 11.0 | 17.0 | 49.0 | 11.0 | 17.0 | 8.0 | 19.0 | 4.0 | 11.0 | 23.0 |
| 04 | 36.0 | 109.0 | 44.0 | 51.0 | 6.0 | 19.0 | 4.0 | 2.0 | 13.0 | 17.0 | 28.0 | 7.0 | 4.0 | 29.0 | 19.0 | 15.0 |
| 05 | 5.0 | 37.0 | 13.0 | 4.0 | 12.0 | 144.0 | 13.0 | 5.0 | 7.0 | 38.0 | 46.0 | 4.0 | 10.0 | 19.0 | 45.0 | 1.0 |
| 06 | 6.0 | 23.0 | 5.0 | 0.0 | 78.0 | 154.0 | 4.0 | 2.0 | 31.0 | 83.0 | 3.0 | 0.0 | 8.0 | 1.0 | 4.0 | 1.0 |
| 07 | 1.0 | 0.0 | 120.0 | 2.0 | 3.0 | 1.0 | 247.0 | 10.0 | 0.0 | 0.0 | 15.0 | 1.0 | 0.0 | 0.0 | 2.0 | 1.0 |
| 08 | 0.0 | 1.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 4.0 | 1.0 | 379.0 | 0.0 | 0.0 | 0.0 | 13.0 | 1.0 |
| 09 | 2.0 | 0.0 | 0.0 | 2.0 | 2.0 | 0.0 | 0.0 | 0.0 | 387.0 | 6.0 | 3.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 10 | 372.0 | 1.0 | 7.0 | 12.0 | 5.0 | 1.0 | 0.0 | 0.0 | 2.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 11 | 45.0 | 16.0 | 315.0 | 5.0 | 0.0 | 1.0 | 2.0 | 0.0 | 1.0 | 2.0 | 4.0 | 0.0 | 0.0 | 4.0 | 8.0 | 0.0 |
| 12 | 12.0 | 5.0 | 0.0 | 29.0 | 5.0 | 10.0 | 0.0 | 8.0 | 39.0 | 74.0 | 23.0 | 193.0 | 1.0 | 1.0 | 2.0 | 1.0 |
| 13 | 8.0 | 10.0 | 34.0 | 5.0 | 19.0 | 29.0 | 31.0 | 11.0 | 8.0 | 7.0 | 10.0 | 0.0 | 35.0 | 41.0 | 126.0 | 29.0 |
| 14 | 21.0 | 7.0 | 36.0 | 6.0 | 12.0 | 28.0 | 31.0 | 16.0 | 35.0 | 57.0 | 83.0 | 26.0 | 6.0 | 5.0 | 27.0 | 7.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.104 | -0.277 | 0.505 | -1.243 | 0.406 | -0.386 | 0.505 | -0.508 | 0.031 | -0.227 | 1.146 | -0.648 | -1.243 | -0.902 | 0.069 | -0.725 |
| 02 | 0.596 | -0.508 | -0.133 | -0.445 | 0.172 | -0.648 | -0.725 | -0.81 | 1.507 | -0.277 | 0.353 | -0.179 | -0.445 | -1.243 | -0.508 | -1.003 |
| 03 | 1.759 | -0.576 | 0.031 | -0.277 | -0.048 | -2.376 | -0.81 | -0.386 | 0.658 | -0.81 | -0.386 | -1.116 | -0.277 | -1.765 | -0.81 | -0.089 |
| 04 | 0.353 | 1.454 | 0.552 | 0.698 | -1.389 | -0.277 | -1.765 | -2.376 | -0.648 | -0.386 | 0.104 | -1.243 | -1.765 | 0.139 | -0.277 | -0.508 |
| 05 | -1.559 | 0.38 | -0.648 | -1.765 | -0.725 | 1.731 | -0.648 | -1.559 | -1.243 | 0.406 | 0.596 | -1.765 | -0.902 | -0.277 | 0.574 | -2.923 |
| 06 | -1.389 | -0.089 | -1.559 | -4.222 | 1.12 | 1.798 | -1.765 | -2.376 | 0.205 | 1.182 | -2.025 | -4.222 | -1.116 | -2.923 | -1.765 | -2.923 |
| 07 | -2.923 | -4.222 | 1.549 | -2.376 | -2.025 | -2.923 | 2.27 | -0.902 | -4.222 | -4.222 | -0.508 | -2.923 | -4.222 | -4.222 | -2.376 | -2.923 |
| 08 | -4.222 | -2.923 | -2.025 | -4.222 | -4.222 | -4.222 | -2.923 | -4.222 | -1.765 | -2.923 | 2.697 | -4.222 | -4.222 | -4.222 | -0.648 | -2.923 |
| 09 | -2.376 | -4.222 | -4.222 | -2.376 | -2.376 | -4.222 | -4.222 | -4.222 | 2.718 | -1.389 | -2.025 | -4.222 | -2.923 | -4.222 | -4.222 | -4.222 |
| 10 | 2.679 | -2.923 | -1.243 | -0.725 | -1.559 | -2.923 | -4.222 | -4.222 | -2.376 | -2.923 | -4.222 | -4.222 | -2.376 | -4.222 | -4.222 | -4.222 |
| 11 | 0.574 | -0.445 | 2.513 | -1.559 | -4.222 | -2.923 | -2.376 | -4.222 | -2.923 | -2.376 | -1.765 | -4.222 | -4.222 | -1.765 | -1.116 | -4.222 |
| 12 | -0.725 | -1.559 | -4.222 | 0.139 | -1.559 | -0.902 | -4.222 | -1.116 | 0.432 | 1.068 | -0.089 | 2.024 | -2.923 | -2.923 | -2.376 | -2.923 |
| 13 | -1.116 | -0.902 | 0.296 | -1.559 | -0.277 | 0.139 | 0.205 | -0.81 | -1.116 | -1.243 | -0.902 | -4.222 | 0.325 | 0.482 | 1.598 | 0.139 |
| 14 | -0.179 | -1.243 | 0.353 | -1.389 | -0.725 | 0.104 | 0.205 | -0.445 | 0.325 | 0.808 | 1.182 | 0.031 | -1.389 | -1.559 | 0.069 | -1.243 |