Model info
| Transcription factor | ELF5 (GeneCards) | ||||||||
| Model | ELF5_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 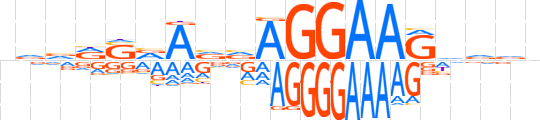 | ||||||||
| LOGO (reverse complement) | 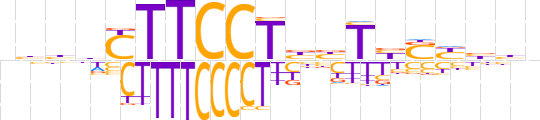 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvvRSvAvvAGGAAGhdvn | ||||||||
| Best auROC (human) | 0.982 | ||||||||
| Best auROC (mouse) | 0.926 | ||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | 12 | ||||||||
| Aligned words | 514 | ||||||||
| TF family | Ets-related factors {3.5.2} | ||||||||
| TF subfamily | EHF-like factors {3.5.2.4} | ||||||||
| HGNC | HGNC:3320 | ||||||||
| EntrezGene | GeneID:2001 (SSTAR profile) | ||||||||
| UniProt ID | ELF5_HUMAN | ||||||||
| UniProt AC | Q9UKW6 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ELF5 expression | ||||||||
| ReMap ChIP-seq dataset list | ELF5 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 63.0 | 15.0 | 76.0 | 13.0 | 32.0 | 26.0 | 22.0 | 15.0 | 55.0 | 25.0 | 48.0 | 11.0 | 19.0 | 20.0 | 46.0 | 12.0 |
| 02 | 71.0 | 30.0 | 61.0 | 7.0 | 45.0 | 18.0 | 5.0 | 18.0 | 86.0 | 26.0 | 72.0 | 8.0 | 7.0 | 2.0 | 32.0 | 10.0 |
| 03 | 32.0 | 12.0 | 158.0 | 7.0 | 35.0 | 5.0 | 18.0 | 18.0 | 63.0 | 19.0 | 78.0 | 10.0 | 9.0 | 4.0 | 25.0 | 5.0 |
| 04 | 13.0 | 17.0 | 107.0 | 2.0 | 6.0 | 13.0 | 13.0 | 8.0 | 32.0 | 30.0 | 205.0 | 12.0 | 9.0 | 6.0 | 21.0 | 4.0 |
| 05 | 26.0 | 16.0 | 15.0 | 3.0 | 32.0 | 24.0 | 5.0 | 5.0 | 203.0 | 37.0 | 93.0 | 13.0 | 12.0 | 6.0 | 4.0 | 4.0 |
| 06 | 242.0 | 3.0 | 24.0 | 4.0 | 71.0 | 1.0 | 3.0 | 8.0 | 103.0 | 2.0 | 7.0 | 5.0 | 20.0 | 0.0 | 3.0 | 2.0 |
| 07 | 94.0 | 88.0 | 225.0 | 29.0 | 4.0 | 1.0 | 1.0 | 0.0 | 9.0 | 4.0 | 23.0 | 1.0 | 3.0 | 6.0 | 8.0 | 2.0 |
| 08 | 16.0 | 27.0 | 64.0 | 3.0 | 52.0 | 30.0 | 13.0 | 4.0 | 86.0 | 47.0 | 113.0 | 11.0 | 12.0 | 5.0 | 14.0 | 1.0 |
| 09 | 144.0 | 5.0 | 17.0 | 0.0 | 102.0 | 5.0 | 1.0 | 1.0 | 186.0 | 3.0 | 15.0 | 0.0 | 14.0 | 1.0 | 4.0 | 0.0 |
| 10 | 0.0 | 0.0 | 443.0 | 3.0 | 2.0 | 0.0 | 9.0 | 3.0 | 0.0 | 0.0 | 36.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 11 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 2.0 | 485.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 |
| 12 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 494.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 488.0 | 2.0 | 1.0 | 6.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 76.0 | 24.0 | 386.0 | 3.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 4.0 | 0.0 | 2.0 | 0.0 |
| 15 | 23.0 | 24.0 | 9.0 | 26.0 | 9.0 | 7.0 | 2.0 | 6.0 | 151.0 | 68.0 | 54.0 | 116.0 | 0.0 | 1.0 | 0.0 | 2.0 |
| 16 | 63.0 | 15.0 | 97.0 | 8.0 | 47.0 | 11.0 | 17.0 | 25.0 | 30.0 | 4.0 | 22.0 | 9.0 | 33.0 | 24.0 | 61.0 | 32.0 |
| 17 | 56.0 | 16.0 | 86.0 | 15.0 | 24.0 | 16.0 | 7.0 | 7.0 | 57.0 | 23.0 | 92.0 | 25.0 | 12.0 | 21.0 | 23.0 | 18.0 |
| 18 | 46.0 | 26.0 | 62.0 | 15.0 | 25.0 | 23.0 | 6.0 | 22.0 | 87.0 | 32.0 | 64.0 | 25.0 | 11.0 | 16.0 | 19.0 | 19.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.699 | -0.717 | 0.885 | -0.856 | 0.027 | -0.177 | -0.342 | -0.717 | 0.564 | -0.216 | 0.429 | -1.018 | -0.486 | -0.435 | 0.387 | -0.934 |
| 02 | 0.818 | -0.036 | 0.667 | -1.451 | 0.365 | -0.539 | -1.766 | -0.539 | 1.008 | -0.177 | 0.832 | -1.324 | -1.451 | -2.58 | 0.027 | -1.11 |
| 03 | 0.027 | -0.934 | 1.615 | -1.451 | 0.116 | -1.766 | -0.539 | -0.539 | 0.699 | -0.486 | 0.911 | -1.11 | -1.211 | -1.971 | -0.216 | -1.766 |
| 04 | -0.856 | -0.595 | 1.226 | -2.58 | -1.596 | -0.856 | -0.856 | -1.324 | 0.027 | -0.036 | 1.874 | -0.934 | -1.211 | -1.596 | -0.388 | -1.971 |
| 05 | -0.177 | -0.654 | -0.717 | -2.23 | 0.027 | -0.256 | -1.766 | -1.766 | 1.865 | 0.171 | 1.086 | -0.856 | -0.934 | -1.596 | -1.971 | -1.971 |
| 06 | 2.04 | -2.23 | -0.256 | -1.971 | 0.818 | -3.122 | -2.23 | -1.324 | 1.188 | -2.58 | -1.451 | -1.766 | -0.435 | -4.397 | -2.23 | -2.58 |
| 07 | 1.097 | 1.031 | 1.967 | -0.07 | -1.971 | -3.122 | -3.122 | -4.397 | -1.211 | -1.971 | -0.298 | -3.122 | -2.23 | -1.596 | -1.324 | -2.58 |
| 08 | -0.654 | -0.14 | 0.715 | -2.23 | 0.508 | -0.036 | -0.856 | -1.971 | 1.008 | 0.408 | 1.28 | -1.018 | -0.934 | -1.766 | -0.784 | -3.122 |
| 09 | 1.522 | -1.766 | -0.595 | -4.397 | 1.178 | -1.766 | -3.122 | -3.122 | 1.777 | -2.23 | -0.717 | -4.397 | -0.784 | -3.122 | -1.971 | -4.397 |
| 10 | -4.397 | -4.397 | 2.644 | -2.23 | -2.58 | -4.397 | -1.211 | -2.23 | -4.397 | -4.397 | 0.144 | -3.122 | -4.397 | -4.397 | -3.122 | -4.397 |
| 11 | -4.397 | -4.397 | -2.58 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -2.58 | -2.58 | 2.735 | -4.397 | -4.397 | -4.397 | -1.451 | -4.397 |
| 12 | -2.58 | -4.397 | -4.397 | -4.397 | -3.122 | -4.397 | -3.122 | -4.397 | 2.753 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 |
| 13 | 2.741 | -2.58 | -3.122 | -1.596 | -4.397 | -4.397 | -4.397 | -4.397 | -3.122 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 |
| 14 | 0.885 | -0.256 | 2.506 | -2.23 | -2.58 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -3.122 | -4.397 | -1.971 | -4.397 | -2.58 | -4.397 |
| 15 | -0.298 | -0.256 | -1.211 | -0.177 | -1.211 | -1.451 | -2.58 | -1.596 | 1.569 | 0.775 | 0.546 | 1.307 | -4.397 | -3.122 | -4.397 | -2.58 |
| 16 | 0.699 | -0.717 | 1.128 | -1.324 | 0.408 | -1.018 | -0.595 | -0.216 | -0.036 | -1.971 | -0.342 | -1.211 | 0.058 | -0.256 | 0.667 | 0.027 |
| 17 | 0.582 | -0.654 | 1.008 | -0.717 | -0.256 | -0.654 | -1.451 | -1.451 | 0.599 | -0.298 | 1.076 | -0.216 | -0.934 | -0.388 | -0.298 | -0.539 |
| 18 | 0.387 | -0.177 | 0.683 | -0.717 | -0.216 | -0.298 | -1.596 | -0.342 | 1.02 | 0.027 | 0.715 | -0.216 | -1.018 | -0.654 | -0.486 | -0.486 |