Model info
| Transcription factor | Esr1 | ||||||||
| Model | ESR1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 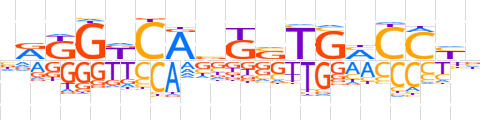 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nRGGKCASCvTGMCMYn | ||||||||
| Best auROC (human) | 0.976 | ||||||||
| Best auROC (mouse) | 0.996 | ||||||||
| Peak sets in benchmark (human) | 432 | ||||||||
| Peak sets in benchmark (mouse) | 58 | ||||||||
| Aligned words | 527 | ||||||||
| TF family | Steroid hormone receptors (NR3) {2.1.1} | ||||||||
| TF subfamily | ER-like receptors (NR3A&B) {2.1.1.2} | ||||||||
| MGI | MGI:1352467 | ||||||||
| EntrezGene | GeneID:13982 (SSTAR profile) | ||||||||
| UniProt ID | ESR1_MOUSE | ||||||||
| UniProt AC | P19785 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Esr1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 52.0 | 10.0 | 52.0 | 5.0 | 123.0 | 8.0 | 16.0 | 19.0 | 79.0 | 6.0 | 30.0 | 5.0 | 21.0 | 8.0 | 57.0 | 8.0 |
| 02 | 11.0 | 4.0 | 236.0 | 24.0 | 3.0 | 0.0 | 18.0 | 11.0 | 7.0 | 0.0 | 121.0 | 27.0 | 3.0 | 0.0 | 31.0 | 3.0 |
| 03 | 0.0 | 0.0 | 24.0 | 0.0 | 2.0 | 1.0 | 1.0 | 0.0 | 33.0 | 15.0 | 344.0 | 14.0 | 2.0 | 1.0 | 62.0 | 0.0 |
| 04 | 9.0 | 3.0 | 15.0 | 10.0 | 7.0 | 1.0 | 0.0 | 9.0 | 52.0 | 18.0 | 81.0 | 280.0 | 1.0 | 1.0 | 3.0 | 9.0 |
| 05 | 3.0 | 56.0 | 7.0 | 3.0 | 0.0 | 21.0 | 0.0 | 2.0 | 0.0 | 91.0 | 1.0 | 7.0 | 3.0 | 244.0 | 44.0 | 17.0 |
| 06 | 3.0 | 0.0 | 3.0 | 0.0 | 371.0 | 5.0 | 21.0 | 15.0 | 35.0 | 3.0 | 13.0 | 1.0 | 16.0 | 2.0 | 10.0 | 1.0 |
| 07 | 27.0 | 242.0 | 117.0 | 39.0 | 1.0 | 5.0 | 0.0 | 4.0 | 4.0 | 28.0 | 7.0 | 8.0 | 0.0 | 12.0 | 2.0 | 3.0 |
| 08 | 10.0 | 22.0 | 0.0 | 0.0 | 132.0 | 155.0 | 0.0 | 0.0 | 41.0 | 85.0 | 0.0 | 0.0 | 7.0 | 47.0 | 0.0 | 0.0 |
| 09 | 27.0 | 58.0 | 100.0 | 5.0 | 70.0 | 149.0 | 33.0 | 57.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 6.0 | 8.0 | 3.0 | 80.0 | 18.0 | 7.0 | 1.0 | 181.0 | 10.0 | 10.0 | 2.0 | 111.0 | 2.0 | 13.0 | 6.0 | 41.0 |
| 11 | 4.0 | 2.0 | 30.0 | 0.0 | 17.0 | 4.0 | 17.0 | 0.0 | 1.0 | 1.0 | 10.0 | 0.0 | 18.0 | 10.0 | 385.0 | 0.0 |
| 12 | 26.0 | 7.0 | 6.0 | 1.0 | 13.0 | 2.0 | 0.0 | 2.0 | 282.0 | 85.0 | 24.0 | 51.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 8.0 | 302.0 | 8.0 | 3.0 | 6.0 | 79.0 | 0.0 | 9.0 | 1.0 | 25.0 | 1.0 | 3.0 | 7.0 | 38.0 | 4.0 | 5.0 |
| 14 | 1.0 | 18.0 | 1.0 | 2.0 | 91.0 | 313.0 | 1.0 | 39.0 | 2.0 | 11.0 | 0.0 | 0.0 | 1.0 | 16.0 | 2.0 | 1.0 |
| 15 | 7.0 | 37.0 | 9.0 | 42.0 | 16.0 | 111.0 | 12.0 | 219.0 | 0.0 | 1.0 | 0.0 | 3.0 | 2.0 | 18.0 | 4.0 | 18.0 |
| 16 | 9.0 | 5.0 | 6.0 | 5.0 | 68.0 | 27.0 | 8.0 | 64.0 | 5.0 | 7.0 | 6.0 | 7.0 | 31.0 | 74.0 | 114.0 | 63.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.506 | -1.112 | 0.506 | -1.768 | 1.363 | -1.326 | -0.656 | -0.488 | 0.922 | -1.598 | -0.038 | -1.768 | -0.39 | -1.326 | 0.597 | -1.326 |
| 02 | -1.02 | -1.973 | 2.013 | -0.258 | -2.232 | -4.398 | -0.541 | -1.02 | -1.452 | -4.398 | 1.347 | -0.142 | -2.232 | -4.398 | -0.006 | -2.232 |
| 03 | -4.398 | -4.398 | -0.258 | -4.398 | -2.582 | -3.124 | -3.124 | -4.398 | 0.056 | -0.719 | 2.389 | -0.786 | -2.582 | -3.124 | 0.681 | -4.398 |
| 04 | -1.213 | -2.232 | -0.719 | -1.112 | -1.452 | -3.124 | -4.398 | -1.213 | 0.506 | -0.541 | 0.947 | 2.184 | -3.124 | -3.124 | -2.232 | -1.213 |
| 05 | -2.232 | 0.58 | -1.452 | -2.232 | -4.398 | -0.39 | -4.398 | -2.582 | -4.398 | 1.063 | -3.124 | -1.452 | -2.232 | 2.046 | 0.341 | -0.597 |
| 06 | -2.232 | -4.398 | -2.232 | -4.398 | 2.465 | -1.768 | -0.39 | -0.719 | 0.114 | -2.232 | -0.858 | -3.124 | -0.656 | -2.582 | -1.112 | -3.124 |
| 07 | -0.142 | 2.038 | 1.313 | 0.221 | -3.124 | -1.768 | -4.398 | -1.973 | -1.973 | -0.106 | -1.452 | -1.326 | -4.398 | -0.936 | -2.582 | -2.232 |
| 08 | -1.112 | -0.344 | -4.398 | -4.398 | 1.433 | 1.594 | -4.398 | -4.398 | 0.271 | 0.995 | -4.398 | -4.398 | -1.452 | 0.406 | -4.398 | -4.398 |
| 09 | -0.142 | 0.615 | 1.157 | -1.768 | 0.802 | 1.554 | 0.056 | 0.597 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 |
| 10 | -1.598 | -1.326 | -2.232 | 0.934 | -0.541 | -1.452 | -3.124 | 1.748 | -1.112 | -1.112 | -2.582 | 1.261 | -2.582 | -0.858 | -1.598 | 0.271 |
| 11 | -1.973 | -2.582 | -0.038 | -4.398 | -0.597 | -1.973 | -0.597 | -4.398 | -3.124 | -3.124 | -1.112 | -4.398 | -0.541 | -1.112 | 2.502 | -4.398 |
| 12 | -0.179 | -1.452 | -1.598 | -3.124 | -0.858 | -2.582 | -4.398 | -2.582 | 2.191 | 0.995 | -0.258 | 0.487 | -4.398 | -4.398 | -4.398 | -4.398 |
| 13 | -1.326 | 2.259 | -1.326 | -2.232 | -1.598 | 0.922 | -4.398 | -1.213 | -3.124 | -0.218 | -3.124 | -2.232 | -1.452 | 0.195 | -1.973 | -1.768 |
| 14 | -3.124 | -0.541 | -3.124 | -2.582 | 1.063 | 2.295 | -3.124 | 0.221 | -2.582 | -1.02 | -4.398 | -4.398 | -3.124 | -0.656 | -2.582 | -3.124 |
| 15 | -1.452 | 0.169 | -1.213 | 0.294 | -0.656 | 1.261 | -0.936 | 1.938 | -4.398 | -3.124 | -4.398 | -2.232 | -2.582 | -0.541 | -1.973 | -0.541 |
| 16 | -1.213 | -1.768 | -1.598 | -1.768 | 0.773 | -0.142 | -1.326 | 0.713 | -1.768 | -1.452 | -1.598 | -1.452 | -0.006 | 0.857 | 1.287 | 0.697 |