Model info
| Transcription factor | Esr2 | ||||||||
| Model | ESR2_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 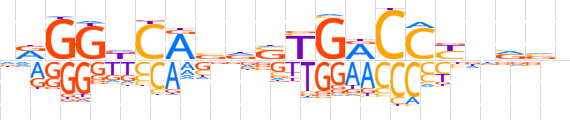 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbbnRGGYCRShvTGhCCYn | ||||||||
| Best auROC (human) | 0.926 | ||||||||
| Best auROC (mouse) | 0.943 | ||||||||
| Peak sets in benchmark (human) | 11 | ||||||||
| Peak sets in benchmark (mouse) | 7 | ||||||||
| Aligned words | 223 | ||||||||
| TF family | Steroid hormone receptors (NR3) {2.1.1} | ||||||||
| TF subfamily | ER-like receptors (NR3A&B) {2.1.1.2} | ||||||||
| MGI | MGI:109392 | ||||||||
| EntrezGene | GeneID:13983 (SSTAR profile) | ||||||||
| UniProt ID | ESR2_MOUSE | ||||||||
| UniProt AC | O08537 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Esr2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.0 | 22.0 | 16.0 | 9.0 | 9.0 | 25.0 | 0.0 | 12.0 | 5.0 | 33.0 | 32.0 | 12.0 | 7.0 | 19.0 | 11.0 | 11.0 |
| 02 | 4.0 | 13.0 | 3.0 | 1.0 | 10.0 | 48.0 | 9.0 | 32.0 | 3.0 | 30.0 | 10.0 | 16.0 | 4.0 | 26.0 | 4.0 | 10.0 |
| 03 | 9.0 | 5.0 | 6.0 | 1.0 | 39.0 | 34.0 | 7.0 | 37.0 | 8.0 | 8.0 | 6.0 | 4.0 | 5.0 | 25.0 | 19.0 | 10.0 |
| 04 | 12.0 | 3.0 | 45.0 | 1.0 | 46.0 | 10.0 | 14.0 | 2.0 | 17.0 | 8.0 | 13.0 | 0.0 | 8.0 | 3.0 | 40.0 | 1.0 |
| 05 | 0.0 | 0.0 | 64.0 | 19.0 | 0.0 | 0.0 | 19.0 | 5.0 | 3.0 | 0.0 | 97.0 | 12.0 | 0.0 | 0.0 | 4.0 | 0.0 |
| 06 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 | 0.0 | 174.0 | 0.0 | 0.0 | 1.0 | 35.0 | 0.0 |
| 07 | 6.0 | 0.0 | 1.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 19.0 | 31.0 | 8.0 | 154.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 23.0 | 2.0 | 0.0 | 0.0 | 23.0 | 1.0 | 7.0 | 0.0 | 9.0 | 0.0 | 0.0 | 0.0 | 149.0 | 7.0 | 2.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 167.0 | 11.0 | 20.0 | 6.0 | 7.0 | 1.0 | 2.0 | 0.0 | 1.0 | 4.0 | 4.0 | 0.0 |
| 10 | 15.0 | 101.0 | 42.0 | 17.0 | 0.0 | 14.0 | 0.0 | 2.0 | 1.0 | 17.0 | 5.0 | 3.0 | 0.0 | 2.0 | 3.0 | 1.0 |
| 11 | 5.0 | 6.0 | 4.0 | 1.0 | 26.0 | 61.0 | 10.0 | 37.0 | 3.0 | 35.0 | 6.0 | 6.0 | 8.0 | 10.0 | 3.0 | 2.0 |
| 12 | 6.0 | 15.0 | 18.0 | 3.0 | 15.0 | 57.0 | 22.0 | 18.0 | 8.0 | 12.0 | 2.0 | 1.0 | 0.0 | 22.0 | 23.0 | 1.0 |
| 13 | 3.0 | 4.0 | 2.0 | 20.0 | 4.0 | 9.0 | 2.0 | 91.0 | 4.0 | 11.0 | 0.0 | 50.0 | 0.0 | 6.0 | 0.0 | 17.0 |
| 14 | 1.0 | 1.0 | 9.0 | 0.0 | 5.0 | 1.0 | 23.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 8.0 | 4.0 | 164.0 | 2.0 |
| 15 | 11.0 | 2.0 | 1.0 | 1.0 | 3.0 | 0.0 | 0.0 | 4.0 | 108.0 | 48.0 | 20.0 | 21.0 | 1.0 | 2.0 | 0.0 | 1.0 |
| 16 | 8.0 | 114.0 | 1.0 | 0.0 | 2.0 | 45.0 | 0.0 | 5.0 | 3.0 | 16.0 | 1.0 | 1.0 | 2.0 | 18.0 | 2.0 | 5.0 |
| 17 | 1.0 | 14.0 | 0.0 | 0.0 | 9.0 | 176.0 | 1.0 | 7.0 | 0.0 | 4.0 | 0.0 | 0.0 | 1.0 | 9.0 | 1.0 | 0.0 |
| 18 | 1.0 | 6.0 | 0.0 | 4.0 | 16.0 | 68.0 | 5.0 | 114.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 4.0 | 3.0 | 0.0 |
| 19 | 4.0 | 3.0 | 10.0 | 0.0 | 24.0 | 18.0 | 16.0 | 20.0 | 0.0 | 2.0 | 6.0 | 2.0 | 11.0 | 34.0 | 35.0 | 38.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -3.743 | 0.448 | 0.135 | -0.424 | -0.424 | 0.574 | -3.743 | -0.146 | -0.984 | 0.848 | 0.818 | -0.146 | -0.665 | 0.304 | -0.23 | -0.23 |
| 02 | -1.191 | -0.068 | -1.453 | -2.367 | -0.323 | 1.22 | -0.424 | 0.818 | -1.453 | 0.754 | -0.323 | 0.135 | -1.191 | 0.612 | -1.191 | -0.323 |
| 03 | -0.424 | -0.984 | -0.812 | -2.367 | 1.014 | 0.878 | -0.665 | 0.961 | -0.538 | -0.538 | -0.812 | -1.191 | -0.984 | 0.574 | 0.304 | -0.323 |
| 04 | -0.146 | -1.453 | 1.156 | -2.367 | 1.177 | -0.323 | 0.004 | -1.809 | 0.194 | -0.538 | -0.068 | -3.743 | -0.538 | -1.453 | 1.039 | -2.367 |
| 05 | -3.743 | -3.743 | 1.506 | 0.304 | -3.743 | -3.743 | 0.304 | -0.984 | -1.453 | -3.743 | 1.92 | -0.146 | -3.743 | -3.743 | -1.191 | -3.743 |
| 06 | -3.743 | -3.743 | -1.453 | -3.743 | -3.743 | -3.743 | -3.743 | -3.743 | -0.323 | -3.743 | 2.502 | -3.743 | -3.743 | -2.367 | 0.906 | -3.743 |
| 07 | -0.812 | -3.743 | -2.367 | -1.453 | -3.743 | -3.743 | -3.743 | -2.367 | 0.304 | 0.786 | -0.538 | 2.381 | -3.743 | -3.743 | -3.743 | -3.743 |
| 08 | -3.743 | 0.492 | -1.809 | -3.743 | -3.743 | 0.492 | -2.367 | -0.665 | -3.743 | -0.424 | -3.743 | -3.743 | -3.743 | 2.348 | -0.665 | -1.809 |
| 09 | -3.743 | -3.743 | -3.743 | -3.743 | 2.461 | -0.23 | 0.354 | -0.812 | -0.665 | -2.367 | -1.809 | -3.743 | -2.367 | -1.191 | -1.191 | -3.743 |
| 10 | 0.072 | 1.96 | 1.087 | 0.194 | -3.743 | 0.004 | -3.743 | -1.809 | -2.367 | 0.194 | -0.984 | -1.453 | -3.743 | -1.809 | -1.453 | -2.367 |
| 11 | -0.984 | -0.812 | -1.191 | -2.367 | 0.612 | 1.458 | -0.323 | 0.961 | -1.453 | 0.906 | -0.812 | -0.812 | -0.538 | -0.323 | -1.453 | -1.809 |
| 12 | -0.812 | 0.072 | 0.25 | -1.453 | 0.072 | 1.39 | 0.448 | 0.25 | -0.538 | -0.146 | -1.809 | -2.367 | -3.743 | 0.448 | 0.492 | -2.367 |
| 13 | -1.453 | -1.191 | -1.809 | 0.354 | -1.191 | -0.424 | -1.809 | 1.856 | -1.191 | -0.23 | -3.743 | 1.26 | -3.743 | -0.812 | -3.743 | 0.194 |
| 14 | -2.367 | -2.367 | -0.424 | -3.743 | -0.984 | -2.367 | 0.492 | -2.367 | -2.367 | -2.367 | -2.367 | -2.367 | -0.538 | -1.191 | 2.443 | -1.809 |
| 15 | -0.23 | -1.809 | -2.367 | -2.367 | -1.453 | -3.743 | -3.743 | -1.191 | 2.027 | 1.22 | 0.354 | 0.402 | -2.367 | -1.809 | -3.743 | -2.367 |
| 16 | -0.538 | 2.081 | -2.367 | -3.743 | -1.809 | 1.156 | -3.743 | -0.984 | -1.453 | 0.135 | -2.367 | -2.367 | -1.809 | 0.25 | -1.809 | -0.984 |
| 17 | -2.367 | 0.004 | -3.743 | -3.743 | -0.424 | 2.514 | -2.367 | -0.665 | -3.743 | -1.191 | -3.743 | -3.743 | -2.367 | -0.424 | -2.367 | -3.743 |
| 18 | -2.367 | -0.812 | -3.743 | -1.191 | 0.135 | 1.566 | -0.984 | 2.081 | -3.743 | -3.743 | -1.809 | -3.743 | -3.743 | -1.191 | -1.453 | -3.743 |
| 19 | -1.191 | -1.453 | -0.323 | -3.743 | 0.533 | 0.25 | 0.135 | 0.354 | -3.743 | -1.809 | -0.812 | -1.809 | -0.23 | 0.878 | 0.906 | 0.988 |