Model info
| Transcription factor | ETV1 (GeneCards) | ||||||||
| Model | ETV1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 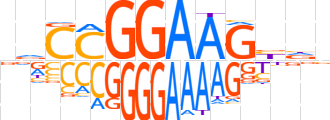 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vdCCGGAAGbdv | ||||||||
| Best auROC (human) | 0.95 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 8 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 499 | ||||||||
| TF family | Ets-related factors {3.5.2} | ||||||||
| TF subfamily | Elk-like factors {3.5.2.2} | ||||||||
| HGNC | HGNC:3490 | ||||||||
| EntrezGene | GeneID:2115 (SSTAR profile) | ||||||||
| UniProt ID | ETV1_HUMAN | ||||||||
| UniProt AC | P50549 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ETV1 expression | ||||||||
| ReMap ChIP-seq dataset list | ETV1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 38.0 | 1.0 | 32.0 | 8.0 | 45.0 | 22.0 | 25.0 | 38.0 | 77.0 | 27.0 | 107.0 | 14.0 | 12.0 | 9.0 | 27.0 | 10.0 |
| 02 | 7.0 | 141.0 | 24.0 | 0.0 | 2.0 | 57.0 | 0.0 | 0.0 | 9.0 | 155.0 | 27.0 | 0.0 | 1.0 | 65.0 | 4.0 | 0.0 |
| 03 | 4.0 | 12.0 | 0.0 | 3.0 | 94.0 | 323.0 | 0.0 | 1.0 | 15.0 | 40.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 04 | 1.0 | 0.0 | 112.0 | 0.0 | 1.0 | 0.0 | 374.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 |
| 05 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 490.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 491.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 446.0 | 0.0 | 1.0 | 44.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 31.0 | 22.0 | 392.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 6.0 | 1.0 | 37.0 | 0.0 |
| 09 | 3.0 | 9.0 | 4.0 | 22.0 | 1.0 | 4.0 | 1.0 | 17.0 | 29.0 | 122.0 | 62.0 | 216.0 | 0.0 | 1.0 | 0.0 | 1.0 |
| 10 | 1.0 | 1.0 | 29.0 | 2.0 | 28.0 | 22.0 | 62.0 | 24.0 | 33.0 | 14.0 | 17.0 | 3.0 | 32.0 | 29.0 | 153.0 | 42.0 |
| 11 | 31.0 | 14.0 | 39.0 | 10.0 | 11.0 | 19.0 | 27.0 | 9.0 | 67.0 | 83.0 | 82.0 | 29.0 | 7.0 | 24.0 | 28.0 | 12.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.209 | -3.111 | 0.039 | -1.312 | 0.377 | -0.33 | -0.204 | 0.209 | 0.91 | -0.128 | 1.238 | -0.772 | -0.922 | -1.199 | -0.128 | -1.098 |
| 02 | -1.439 | 1.513 | -0.244 | -4.387 | -2.568 | 0.611 | -4.387 | -4.387 | -1.199 | 1.608 | -0.128 | -4.387 | -3.111 | 0.742 | -1.96 | -4.387 |
| 03 | -1.96 | -0.922 | -4.387 | -2.218 | 1.109 | 2.34 | -4.387 | -3.111 | -0.705 | 0.26 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 |
| 04 | -3.111 | -4.387 | 1.284 | -4.387 | -3.111 | -4.387 | 2.487 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -1.96 | -4.387 |
| 05 | -4.387 | -4.387 | -2.568 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | 2.757 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 |
| 06 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | 2.759 | -4.387 | -3.111 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 |
| 07 | 2.663 | -4.387 | -3.111 | 0.355 | -4.387 | -4.387 | -4.387 | -4.387 | -3.111 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 |
| 08 | 0.008 | -0.33 | 2.534 | -2.568 | -4.387 | -4.387 | -4.387 | -4.387 | -3.111 | -4.387 | -4.387 | -4.387 | -1.584 | -3.111 | 0.183 | -4.387 |
| 09 | -2.218 | -1.199 | -1.96 | -0.33 | -3.111 | -1.96 | -3.111 | -0.583 | -0.058 | 1.369 | 0.695 | 1.939 | -4.387 | -3.111 | -4.387 | -3.111 |
| 10 | -3.111 | -3.111 | -0.058 | -2.568 | -0.092 | -0.33 | 0.695 | -0.244 | 0.07 | -0.772 | -0.583 | -2.218 | 0.039 | -0.058 | 1.595 | 0.308 |
| 11 | 0.008 | -0.772 | 0.235 | -1.098 | -1.006 | -0.474 | -0.128 | -1.199 | 0.772 | 0.985 | 0.973 | -0.058 | -1.439 | -0.244 | -0.092 | -0.922 |