Model info
| Transcription factor | Fosl1 | ||||||||
| Model | FOSL1_MOUSE.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 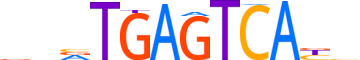 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ddvTGAGTCAbh | ||||||||
| Best auROC (human) | 0.988 | ||||||||
| Best auROC (mouse) | 0.952 | ||||||||
| Peak sets in benchmark (human) | 21 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Fos-related factors {1.1.2} | ||||||||
| TF subfamily | Fos factors {1.1.2.1} | ||||||||
| MGI | MGI:107179 | ||||||||
| EntrezGene | GeneID:14283 (SSTAR profile) | ||||||||
| UniProt ID | FOSL1_MOUSE | ||||||||
| UniProt AC | P48755 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Fosl1 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 99.0 | 93.0 | 203.0 | 105.0 |
| 02 | 159.0 | 82.0 | 174.0 | 85.0 |
| 03 | 215.0 | 135.0 | 141.0 | 9.0 |
| 04 | 0.0 | 0.0 | 3.0 | 497.0 |
| 05 | 0.0 | 2.0 | 471.0 | 27.0 |
| 06 | 494.0 | 1.0 | 1.0 | 4.0 |
| 07 | 39.0 | 0.0 | 461.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 500.0 |
| 09 | 9.0 | 490.0 | 1.0 | 0.0 |
| 10 | 498.0 | 2.0 | 0.0 | 0.0 |
| 11 | 11.0 | 206.0 | 81.0 | 202.0 |
| 12 | 105.0 | 201.0 | 77.0 | 117.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.23 | -0.291 | 0.48 | -0.172 |
| 02 | 0.238 | -0.415 | 0.327 | -0.38 |
| 03 | 0.537 | 0.076 | 0.119 | -2.484 |
| 04 | -4.4 | -4.4 | -3.325 | 1.371 |
| 05 | -4.4 | -3.573 | 1.317 | -1.489 |
| 06 | 1.365 | -3.903 | -3.903 | -3.126 |
| 07 | -1.138 | -4.4 | 1.296 | -4.4 |
| 08 | -4.4 | -4.4 | -4.4 | 1.377 |
| 09 | -2.484 | 1.357 | -3.903 | -4.4 |
| 10 | 1.373 | -3.573 | -4.4 | -4.4 |
| 11 | -2.311 | 0.495 | -0.427 | 0.475 |
| 12 | -0.172 | 0.47 | -0.477 | -0.065 |