Model info
| Transcription factor | FOS (GeneCards) | ||||||||
| Model | FOS_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 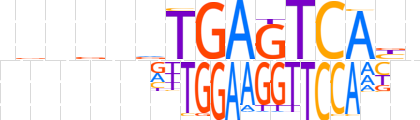 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvnbnvTGAGTCAbn | ||||||||
| Best auROC (human) | 0.991 | ||||||||
| Best auROC (mouse) | 0.92 | ||||||||
| Peak sets in benchmark (human) | 38 | ||||||||
| Peak sets in benchmark (mouse) | 50 | ||||||||
| Aligned words | 363 | ||||||||
| TF family | Fos-related factors {1.1.2} | ||||||||
| TF subfamily | Fos factors {1.1.2.1} | ||||||||
| HGNC | HGNC:3796 | ||||||||
| EntrezGene | GeneID:2353 (SSTAR profile) | ||||||||
| UniProt ID | FOS_HUMAN | ||||||||
| UniProt AC | P01100 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | FOS expression | ||||||||
| ReMap ChIP-seq dataset list | FOS datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 32.0 | 19.0 | 28.0 | 12.0 | 32.0 | 27.0 | 19.0 | 11.0 | 15.0 | 26.0 | 28.0 | 4.0 | 15.0 | 22.0 | 51.0 | 7.0 |
| 02 | 30.0 | 13.0 | 36.0 | 15.0 | 49.0 | 15.0 | 10.0 | 20.0 | 27.0 | 27.0 | 43.0 | 29.0 | 4.0 | 9.0 | 15.0 | 6.0 |
| 03 | 22.0 | 24.0 | 52.0 | 12.0 | 14.0 | 13.0 | 10.0 | 27.0 | 13.0 | 23.0 | 53.0 | 15.0 | 2.0 | 16.0 | 36.0 | 16.0 |
| 04 | 18.0 | 10.0 | 17.0 | 6.0 | 32.0 | 9.0 | 8.0 | 27.0 | 34.0 | 34.0 | 33.0 | 50.0 | 11.0 | 8.0 | 31.0 | 20.0 |
| 05 | 25.0 | 12.0 | 54.0 | 4.0 | 34.0 | 12.0 | 14.0 | 1.0 | 37.0 | 26.0 | 19.0 | 7.0 | 17.0 | 23.0 | 42.0 | 21.0 |
| 06 | 0.0 | 1.0 | 0.0 | 112.0 | 0.0 | 3.0 | 0.0 | 70.0 | 0.0 | 0.0 | 0.0 | 129.0 | 0.0 | 15.0 | 0.0 | 18.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 18.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 327.0 | 2.0 |
| 08 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 345.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 09 | 7.0 | 0.0 | 300.0 | 41.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 300.0 | 0.0 | 0.0 | 0.0 | 41.0 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 346.0 | 0.0 | 0.0 |
| 12 | 2.0 | 0.0 | 0.0 | 0.0 | 329.0 | 14.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 6.0 | 139.0 | 71.0 | 115.0 | 0.0 | 14.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 1.0 | 0.0 |
| 14 | 2.0 | 3.0 | 1.0 | 0.0 | 44.0 | 42.0 | 19.0 | 50.0 | 27.0 | 19.0 | 12.0 | 14.0 | 21.0 | 45.0 | 24.0 | 25.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.381 | -0.133 | 0.249 | -0.581 | 0.381 | 0.213 | -0.133 | -0.666 | -0.364 | 0.176 | 0.249 | -1.622 | -0.364 | 0.011 | 0.843 | -1.099 |
| 02 | 0.317 | -0.504 | 0.497 | -0.364 | 0.803 | -0.364 | -0.758 | -0.082 | 0.213 | 0.213 | 0.673 | 0.284 | -1.622 | -0.859 | -0.364 | -1.245 |
| 03 | 0.011 | 0.097 | 0.862 | -0.581 | -0.431 | -0.504 | -0.758 | 0.213 | -0.504 | 0.055 | 0.881 | -0.364 | -2.235 | -0.301 | 0.497 | -0.301 |
| 04 | -0.186 | -0.758 | -0.242 | -1.245 | 0.381 | -0.859 | -0.972 | 0.213 | 0.441 | 0.441 | 0.411 | 0.823 | -0.666 | -0.972 | 0.349 | -0.082 |
| 05 | 0.137 | -0.581 | 0.899 | -1.622 | 0.441 | -0.581 | -0.431 | -2.785 | 0.524 | 0.176 | -0.133 | -1.099 | -0.242 | 0.055 | 0.65 | -0.035 |
| 06 | -4.102 | -2.785 | -4.102 | 1.625 | -4.102 | -1.883 | -4.102 | 1.157 | -4.102 | -4.102 | -4.102 | 1.766 | -4.102 | -0.364 | -4.102 | -0.186 |
| 07 | -4.102 | -4.102 | -4.102 | -4.102 | -2.785 | -4.102 | -0.186 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | 2.695 | -2.235 |
| 08 | -2.785 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | 2.748 | -4.102 | -4.102 | -4.102 | -2.235 | -4.102 | -4.102 | -4.102 |
| 09 | -1.099 | -4.102 | 2.609 | 0.626 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 |
| 10 | -4.102 | -4.102 | -4.102 | -1.099 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | 2.609 | -4.102 | -4.102 | -4.102 | 0.626 |
| 11 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -2.235 | 2.751 | -4.102 | -4.102 |
| 12 | -2.235 | -4.102 | -4.102 | -4.102 | 2.701 | -0.431 | -4.102 | -1.883 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 |
| 13 | -1.245 | 1.841 | 1.172 | 1.652 | -4.102 | -0.431 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -4.102 | -2.235 | -2.785 | -4.102 |
| 14 | -2.235 | -1.883 | -2.785 | -4.102 | 0.696 | 0.65 | -0.133 | 0.823 | 0.213 | -0.133 | -0.581 | -0.431 | -0.035 | 0.718 | 0.097 | 0.137 |