Model info
| Transcription factor | FOXD3 (GeneCards) | ||||||||
| Model | FOXD3_HUMAN.H11MO.0.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 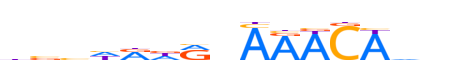 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | D | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bbTGTTTnYhYddbh | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.786 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 11 | ||||||||
| Aligned words | 465 | ||||||||
| TF family | Forkhead box (FOX) factors {3.3.1} | ||||||||
| TF subfamily | FOXD {3.3.1.4} | ||||||||
| HGNC | HGNC:3804 | ||||||||
| EntrezGene | GeneID:27022 (SSTAR profile) | ||||||||
| UniProt ID | FOXD3_HUMAN | ||||||||
| UniProt AC | Q9UJU5 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | FOXD3 expression | ||||||||
| ReMap ChIP-seq dataset list | FOXD3 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 73.0 | 82.0 | 139.0 | 171.0 |
| 02 | 55.0 | 101.0 | 124.0 | 185.0 |
| 03 | 63.0 | 3.0 | 26.0 | 373.0 |
| 04 | 18.0 | 32.0 | 406.0 | 9.0 |
| 05 | 43.0 | 24.0 | 17.0 | 381.0 |
| 06 | 17.0 | 52.0 | 23.0 | 373.0 |
| 07 | 25.0 | 0.0 | 44.0 | 396.0 |
| 08 | 158.0 | 93.0 | 105.0 | 109.0 |
| 09 | 27.0 | 279.0 | 29.0 | 130.0 |
| 10 | 137.0 | 94.0 | 19.0 | 215.0 |
| 11 | 51.0 | 104.0 | 33.0 | 277.0 |
| 12 | 188.0 | 22.0 | 68.0 | 187.0 |
| 13 | 107.0 | 66.0 | 199.0 | 93.0 |
| 14 | 47.0 | 169.0 | 162.0 | 87.0 |
| 15 | 166.0 | 136.0 | 52.0 | 111.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.458 | -0.344 | 0.177 | 0.382 |
| 02 | -0.734 | -0.139 | 0.064 | 0.46 |
| 03 | -0.602 | -3.257 | -1.453 | 1.157 |
| 04 | -1.797 | -1.256 | 1.241 | -2.414 |
| 05 | -0.973 | -1.529 | -1.849 | 1.178 |
| 06 | -1.849 | -0.789 | -1.569 | 1.157 |
| 07 | -1.49 | -4.34 | -0.95 | 1.216 |
| 08 | 0.303 | -0.22 | -0.1 | -0.064 |
| 09 | -1.418 | 0.868 | -1.35 | 0.11 |
| 10 | 0.162 | -0.209 | -1.747 | 0.609 |
| 11 | -0.807 | -0.11 | -1.227 | 0.861 |
| 12 | 0.476 | -1.61 | -0.527 | 0.47 |
| 13 | -0.082 | -0.556 | 0.532 | -0.22 |
| 14 | -0.887 | 0.37 | 0.328 | -0.285 |
| 15 | 0.352 | 0.155 | -0.789 | -0.046 |